Fig. 1.
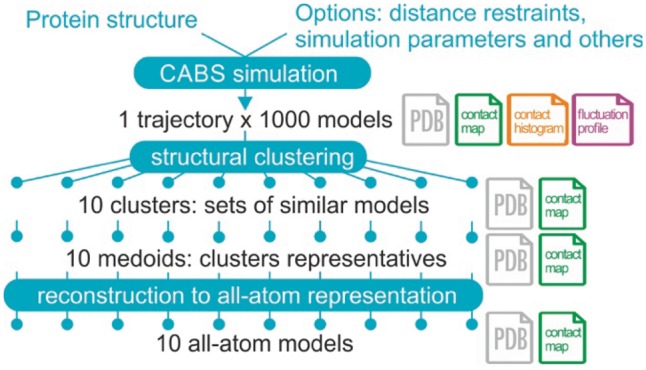
Pipeline of CABS-flex standalone in the default mode. CABS-flex combines CABS coarse-grained model with structural clustering of the simulation results and reconstruction of selected models to all-atom representation. Number of models generated at each modeling step can be set up by the user. In the default mode, CABS-flex trajectory counts 1000 models in coarse-grained representation that are clustered to 10 representative models, subsequently reconstructed to all-atom models. In addition to protein models in PDB format, CABS-flex standalone provides a number of analysis and visualization options, such as contact maps and histograms (marked in the figure, in the appropriate pipeline stages)
