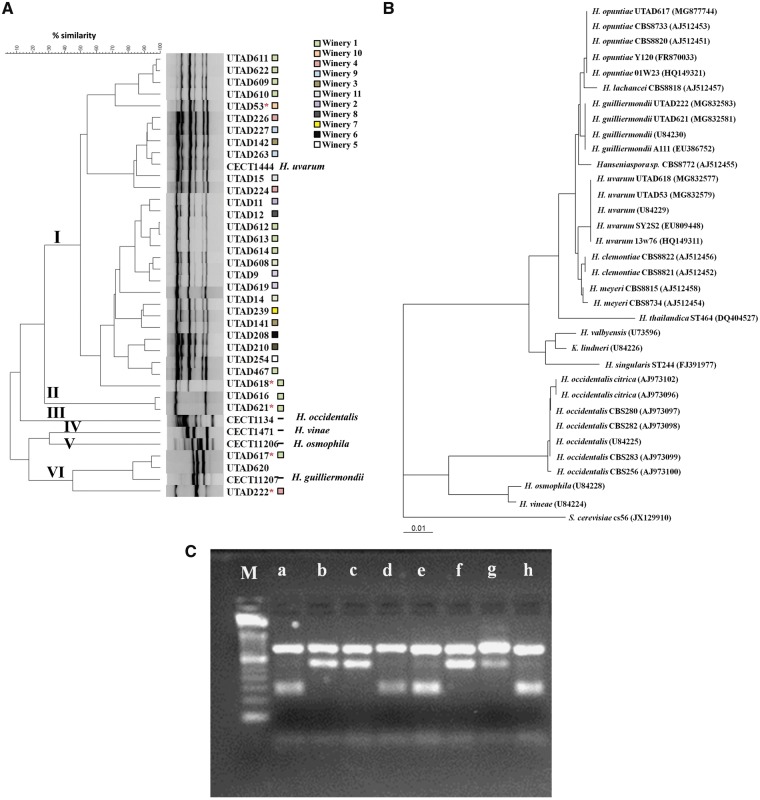
Figure 1.
Genetic characterization, based on PCR-fingerprinting, of the 33 UTAD Hanseniaspora strains recovered from the Douro demarcated region and of the selected reference strains H. guilliermondii CECT11027T, H. occidentalis CECT11341T, H. osmophila CECT 11206T, H. uvarum CECT1444T and H. vineae CECT1471T; (A) Dendrogram obtained by hierarchical analysis of PCR (GTG)5 patterns using Pearson’s correlation coefficient and UPGMA clustering method; (B) Neighbour-joining phylogenetic tree showing the relationships of selected Hanseniaspora strains (marked with an asterisk in A), inferred from the sequences of the D1/D2 domain of the LSU RNA gene. Bootstrap percentages >50% from 1,000 bootstrap replicates are shown. The outgroup species was Saccharomyces cerevisiae. Bar, 1% sequence divergence; (C) Restriction patterns obtained upon restriction with DraI of the ITS region of Hanseniaspora isolates or references strain: M, molecular marker; (a) isolate UTAD616, (b) isolate UTAD617, (c) isolate UTAD620, (d) isolate UTAD621, (e) isolate UTAD222, (f) reference strain H. guilliermondii CECT11027, (g) type strain H. opuntiae CBS8733T and (h) type strain H. guilliermondii CBS465T. The squares designate the different wineries from where the isolates were retrieved.