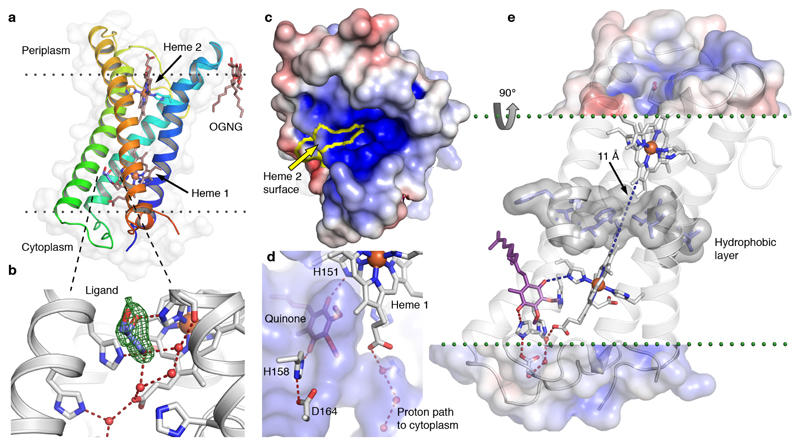
Fig. 3. High-resolution structure of CybB.
(a) 1.97 Å crystal structure of CybB colored from the N-terminus (blue) to the C-terminus (red), the protein surface is indicated (light grey). The calculated location of the hydrophobic core of the membrane is illustrated by dotted lines. (b) Binding site near the cytoplasmic heme, a glycerol molecule was modeled in the cavity based on the electron density, Fo-Fc omit map contoured at 4σ (green mesh). A chain of hydrogen bonded water molecules leads from the ligand-binding site to the cytoplasm. (c) Electrostatic potential map of the protein surface viewed from the periplasm, positive surface potential in blue, negative in red. The molecular surface contributed by the exposed heme 2 porphyrin edge, presumably acting as an electron sink, is bounded in yellow. (d) Protein surface making up the ligand binding cavity and channel to the cytoplasm in blue (viewed from inside the protein). The ubiquinone headgroup (purple) was predicted to occupy the same volume as the bound ligand with its carbonyl groups hydrogen bonded to the iron-coordinating H151 and a second completely conserved histidine (H158). (e) Proposed functional architecture of CybB. The positively charged funnel attracts superoxide to the heme edge, which serves as an electron sink and oxidizes the superoxide to molecular oxygen. The electron is subsequently tunneled to the quinone binding site (blue dashed lines). A layer of hydrophobic residues (gray surface) is located between the hemes, preventing the presence of water molecules or a proton-transfer path through the membrane. Upon quinone reduction, protons are sequestered from the cytoplasm via the indicated hydrogen-bonded path (red) to produce reduced ubiquinol.