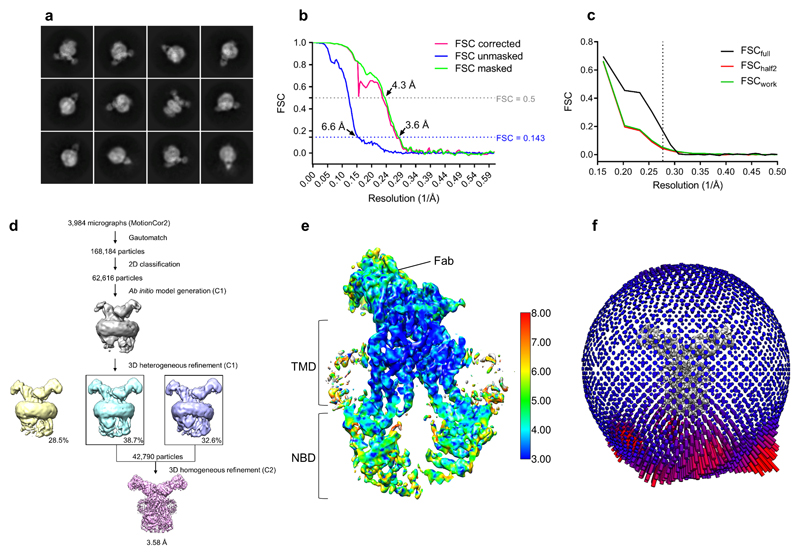
Extended data figure 3. Cryo-EM map generation, data processing and atomic model refinement of ABCG2EQ-E1S.
a, Twelve representative 2D class averages of the final round of 2D classification, sorted in decreasing order by the number of particles assigned to each class. b, FSC from the CryoSPARC auto-refine procedure of the unmasked half-maps (blue), the half-maps after masking (green), and the half-maps after masking and correction for the influence of the mask (pink). A horizontal dotted line (blue) is drawn for the FSC = 0.143 criterion. For both the unmasked and the corrected FSC curves, their intersection with the FSC?=?0.143 line is indicated by an arrow, and the resolution at this point is indicated. c, FSC curve of the final 3.58 Å refined model versus the map it was refined against (FSCfull, black line). FSC curve of the final model with introduced shifts (mean value of 0.3 Å) refined against the first of two independent half-maps (half-map 1) (to which it was refined against; FSCwork green line) or the same refined model versus the second independent half-map (to which is was not refined; FSChalf2, red line). d, The flow chart for the cryo-EM data processing and structure determination of the ABCG2EQ-E1S complex. e, Full view of the final CryoSPARC B-factor sharpened map of ABCG2EQ-E1S coloured by local resolution in Å as calculated by ResMap with the clipping plane in the middle of the molecule. f, Angular distribution plot for the final reconstruction.