Figure 1. KRT9 is a robust marker of volar skin in vivo and is partially maintained in cultured volar keratinocytes in the absence of fibroblasts.
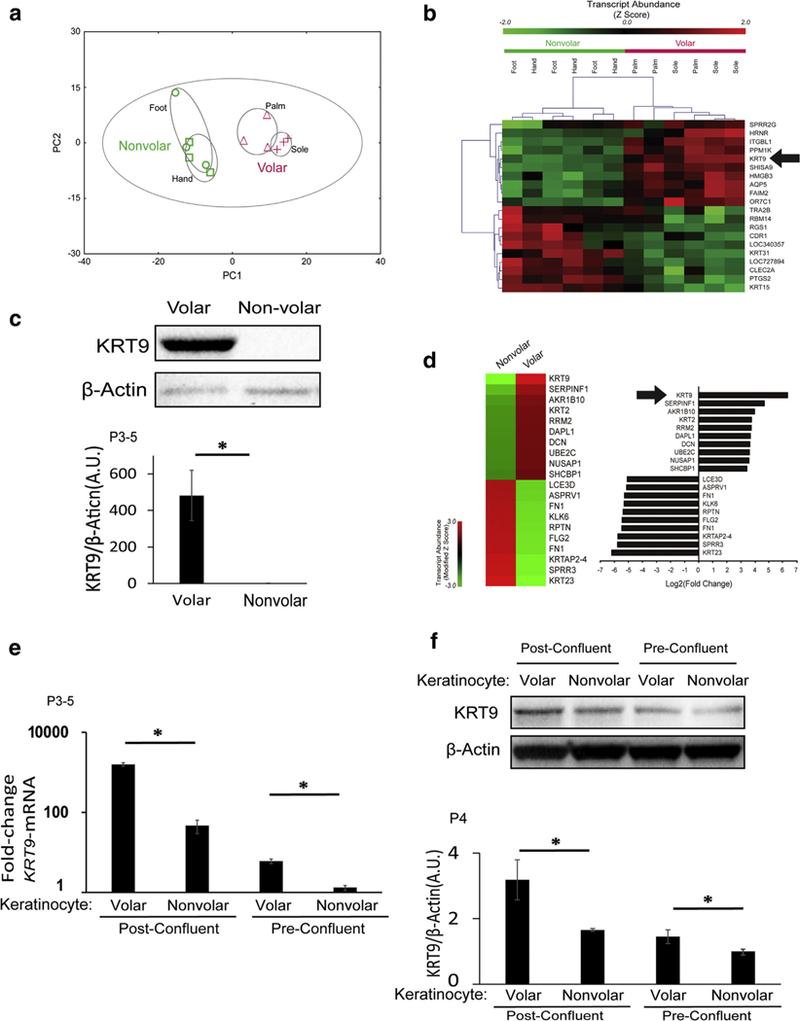
(a) Principle component analysis of microarray data from human volar and nonvolar epidermis show clear clustering of volar skin tissue. n = 3 samples from each site. (b) Unsupervised clustering of expression data from human volar and nonvolar tissue shows that KRT9 is one of the most unique genes in volar skin. n = 3 samples from each site). KRT9 had the highest fold change transcript in volar tissue (30.6-fold, P = 0.0002) (Kim et al., 2016 under GEO: GSE39267. (c) KRT9 protein expression is robust in volar compared with nonvolar human epidermis, as measured by Western blot (top) and normalized to b-Actin (bottom). n = 3, *P < 0.05. (d) In early passaged in vitro solo culture of keratinocytes, microarray analysis shows that KRT9 is maintained as the most unique transcript in volar keratinocytes compared with nonvolar keratinocytes, under GEO: GSE116055. (e) KRT9 mRNA expression of volar keratinocytes is higher than nonvolar keratinocytes both in preconfluent and postconfluent cultures, as measured by quantitative real-time reverse transcriptase–PCR and normalized to RPLP0. n = 3, *P < 0.05. (f) KRT9 protein expression is still elevated in volar compared with nonvolar keratinocytes in both postconfluent and preconfluent conditions, as measured by Western blot (top) and normalized to b-Actin (bottom). n = 3, *P < 0.05. A.U., arbitrary unit; GEO, Gene Expression Omnibus; KRT, Keratin; P, passage.
