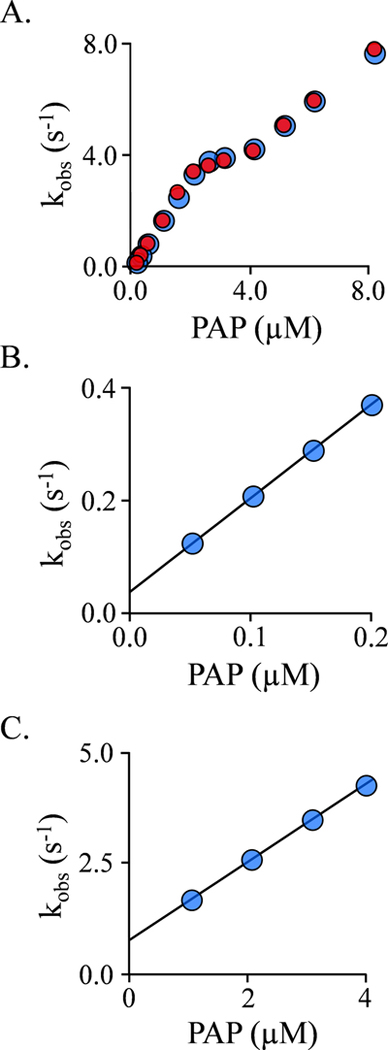
Figure 6. Presteady State binding of PAP to SULT1A1(EGCG)2.
A.) kobs vs [PAP]. The binding- reaction progress curves were monitored using a stopped-flow fluorimeter (λex = 290 nm, λem ≥ 325 nm (cutoff filter). kobs values (black dots) were obtained by fitting 6–9 averaged progress curves using a single-exponential equation. Each kobs value is the average of three independent determinations. Red dots indicate kobs values predicted using the kon and koff values obtained from the experiments associated with Panels B and C. B.) Binding at the High-Affinity Site (kobs vs [PAP]). Reactions were initiated by mixing (1:1) a solution containing SULT1A1 (75 nM, dimer), EGCG (12 μM, 18 ▪ Kd cap open form), MgCh2 (5.0 mM), NaPO4 (50 mM), pH 7.5, 25 ± 2°C, with a solution that was identical except that it lacked SULT1A1 and contained PAP at the indicated concentrations. C.) Binding at the Low-Affinity Site (kobs vs [PAP]). Reactions were initiated by rapidly mixing (1:1) a solution containing SULT1A1 (1.0 μM, dimer), 1.0 μM PAP (42 ▪ Kd high affinity and 1.3 ▪ Kd low affmity), EGCG (12 μm, 18 ▪ Kd cap open form), MgCl2 (5.0 mM), NaPO4 (50 mM), pH 7.5, 25 ± 2°C, with a solution that was identical except that it lacked SULT1A1 and contained PAP at the indicated concentrations. Pre-equilibration at [PAP] = [SULT1A1]dimer saturates the high-affinity nucleotide-binding site and thus prevents it from contributing to the low-affinity binding measurements.