Abstract
Disulfiram (DSF) was well tolerated and activated viral transcription (cell-associated unspliced [CA-US] and plasma HIV RNA) in a Phase 2 dose-escalation trial in HIV+ antiretroviral therapy (ART)-suppressed participants. Here, we investigated whether exposure to disulfiram and its metabolites predicted these changes in HIV transcription. Participants were administered 500 (N=10), 1000 (N=10), or 2000 (N=10) mg of DSF for 3 consecutive days. Disulfiram and four metabolites were measured by ultra-performance liquid chromatography–tandem mass spectrometry (UPLC-MS/MS). Changes in CA-US and plasma HIV RNA were quantified by PCR and analyzed in NONMEM. A seven-compartment pharmacokinetic model demonstrated non-linear elimination kinetics. The fitted median area under the curve values (AUC0–72) were 3816, 8386, and 22331 mg*hr/L, respectively. Higher exposure predicted greater increases in CA-US (Emax=78%, AUC50=1600 mcg*hr/L, P=0.013) but not plasma HIV RNA. These results provide support for further development of disulfiram as an important drug for future HIV cure strategies.
Keywords: pharmacokinetics-pharmacodynamics, HIV, clinical trials, translational medicine
INTRODUCTION
During effective antiretroviral therapy (ART) suppression, HIV persists in latently-infected cells, “the HIV reservoir.” Strategies to eradicate HIV include “shock and kill” approaches that aim to reactivate latently-infected cells to reduce the size of the overall HIV reservoir (1). Reduction of the HIV reservoir may then lead to a “functional” cure, allowing HIV-infected individuals to maintain host-mediated viral control in the absence of ART. Disulfiram (Antabuse®), a drug used to treat alcoholism, was identified from a large screen of a library of FDA-approved compounds that could induce in vitro HIV gene expression using a model for latent HIV infection (2, 3). These results provided an intriguing hypothesis as to whether disulfiram (DSF) could potentially be used in future HIV eradication strategies (4). In vitro experiments showed that disulfiram and its metabolite, diethyldithiocarbamate methyl ester (DDTC-Me), activates latent HIV in Bcl-2-transduced primary CD4+ T cells (5). In a subsequent Phase 1 trial, DSF 500 mg administered to HIV+ antiretroviral therapy (ART)-suppressed participants for 14 days led to transient but non-statistically significant increases in plasma HIV RNA, primarily among participants with higher plasma concentrations of disulfiram (6). DSF administration also resulted in considerable inter-individual plasma DSF pharmacokinetic (PK) variability, consistent with prior reports in alcoholic participants receiving DSF (7).
Quantifying the “HIV reservoir” is challenging – current methods may either underestimate or overestimate the total number of HIV-infected cells harboring replication-competent virus (8). Therefore, a major challenge in performing clinical trials is how best to assess the efficacy of HIV latency reveral agents and determine a true drug exposure-response. The most sensitive assays to measure HIV-infected cells are polymerase chain reaction (PCR)-based assays quantifying cell-associated [CA] HIV RNA or DNA. Measures of CA-RNA include unspliced (CA-US, translated into structural HIV proteins Pol and Gag) and multi-spliced (CA-ms, reflective of the ability of the cell to produce virus) RNA (9, 10). In contrast, ultra-sensitive assays to measure plasma (i.e., “cell-free”) viremia can detect virion-associated HIV RNA at the single copy level but do not necessarily represent the total replication-competent reservoir (11). Culture-based assays to quantify the outgrowth of replication-competent virus are expensive, laborious, and underestimate the size of the inducible reservoir (~300-fold less than CA DNA) (12), precluding its widespread utility in estimating potential small effects from these early HIV latency reversal agent trials.
Disulfiram was first described in 1937 as a potential therapeutic agent for alcoholism after rubber factory workers exposed to disulfiram demonstrated symptoms of nausea, vomiting, flushing, palpitations, headache, and circulatory changes, which were later discovered to be due to the accumulation of acetaldehyde (13). After oral administration, disulfiram is 80–95% absorbed from the gastrointestinal tract and is distributed widely into tissues – with the highest concentrations reported in the kidney, pancreas, liver, and gastrointestinal tract and lowest concentrations in the brain (14). DSF in the gut is rapidly reduced to diethyldithiocarbamate (DDTC) and then spontaneously converted to carbon disulfide (CS) and diethylamine (DEA) or methylated in the liver and kidneys to form DDTC-Me. DDTC-Me is then bioactivated primarily via cytochrome P450 (CYP) 3A4/5 to S-methyl-N,N-diethylthiolcarbamate sulfoxide (DETC-MeSO) (15) and eventually metabolized to carbamathione (16). The estimated half-life of disulfiram in humans is about 7 hours, with over 90% of DSF eliminated within 3 days and approximately 7% of disulfiram excreted in feces and 12% by breath as CS (17). However, the rate at which disulfiram is metabolized varies considerably between individuals (7, 18).
Therefore, in desiging the subsequent Phase 2 dose-escalation study evaluating 500, 1000, or 2000 mg daily of disulfiram in HIV+ ART-suppressed participants, we included measurement of DSF and four metabolites to more accurately quantify the exposure to DSF and focused on three well-validated measures of the HIV reservoir: cell-associated unspliced (CA-US) HIV RNA from CD4+ T cells, cell-associated HIV DNA from CD4+ T cells, and plasma HIV RNA (using an ultra-sensitive assay with a lower limit of detection of 1 copy of HIV RNA per mL). In this study, three consecutive days of DSF at all doses led to statistically significant dose-dependent increases in CA-US RNA, but no significant differences were observed in CA-DNA (19). Plasma HIV RNA was also increased, but only at day 31 and only in participants receiving the highest dose (2000 mg) of disulfiram.
Here, in order to comprehensively study the effects of DSF on latent HIV transcription, we performed population pharmacokinetic/pharmacodynamic (PK/PD) modeling using plasma measurements of DSF and four of its metabolites in relation to CA-US and plasma HIV RNA from this Phase 2 study (19). DSF and its metabolites demonstrated non-linear elimination kinetics (greater-than-dose-proportional increases in plasma concentrations for participants in the 2000 mg group). We also observed a statistically significant exposure-response association with CA-US HIV RNA. Exposure-response relationships were less clear for plasma HIV RNA; participants with the higher baseline plasma HIV RNA demonstrated greater increases post-DSF but several of these were in the lowest dosing group potentially obscuring a true exposure-response relationship.
RESULTS
Baseline characteristics of the 30 HIV+ ART-suppressed participants enrolled in the Phase 2 trial were overall balanced between the three dosing groups (Table 1). Participants were mostly male with a median age of 54 years and with median pre-ART nadir CD4+ T cell count=149 cells/mm3 and plasma HIV RNA by routine clinical assay=4.8 log10copies/mL (TaqMan version 2, Roche; NJ, USA; lower limit of detection [LLOD] of 20 copies/mL). Mean baseline values for the HIV reservoir measures, CA-US HIV RNA (LLOD=1 copy equivalent/reaction) and plasma HIV RNA (by ultra-sensitive hybrid real time/digital PCR assay; LLOD 0.19 copies/mL) were not statistically different between dosing cohorts, though there was a trend toward higher baseline plasma HIV RNA in the lowest dose 500 mg cohort (Table 1). Of note, the final LLOD for CA-US RNA and plasma HIV RNA was dependent on the input cell number and volume, respectively. All participants had initiated ART during chronic HIV infection and were ART-suppressed for a median of 8 years. Current ART drug class regimens were similarly distributed among the three dosing cohorts.
Table 1.
Baseline characteristics of HIV-infected antiretroviral therapy (ART)-suppressed study participants.
| Disulfiram Dose Cohort Characteristics |
500 mg N = 10 |
1000 mg N = 10 |
2000 mg N = 10 |
Total N = 30 |
|---|---|---|---|---|
| Gender (male vs. MTF transgender) a | 9 (90%) | 9 (90%) | 10 (100%) | 28 (98%) |
| Age (years) b | 54 (34 – 63) | 54 (40 – 65) | 54 (26 – 67) | 54 (26 – 67) |
| Pre-ART HIV RNA (log10 copies/mL) b | 4.2 (3.0 – 5.4) | 4.9 (4.7 – 5.9) | 4.8 (4.2 – 5.6) | 4.8 (3.0 – 5.9) |
| Nadir CD4+ T cell count (cells/mm3) b | 182 (2 – 380) | 96 (7 – 680) | 132 (1 – 666) | 149 (1 – 680) |
| Duration of ART (years) b | 5 (3 – 15) | 13 (3 – 29) | 10 (2 – 19) | 8 (2 – 29) |
| Proximal CD4+ T cell count (cells/mm3) b, c | 560 (401 – 1180) | 610 (390 – 1137) | 527 (414 – 1022) | 582 (390 – 1180) |
| Antiretroviral regimen (non-NRTI drug) a | ||||
| PI | 5 (50%) | 4 (40%) | 3 (30%) | 12 (40%) |
| NNRTI | 5 (50%) | 4 (40%) | 6 (60%) | 15 (50%) |
| INI | 0 | 2 (20%) | 1 (10%) | 3 (10%) |
| CA-US HIV RNA (copies/106 CD4+ T cells) b, d | 99 (2.2–557) | 69 (6.7–4111) | 113 (9.7–1598) | 99 (2.2–4111) |
| Plasma HIV RNA (copies/mL) b, d | 5.7 (0–25) | 2.6 (0–10) | 0.87 (0–12) | 2.3 (0–25) |
Abbreviations: MTF = male-to-female; NRTI = nucleoside reverse transcriptase inhibitor; PI = protease inhibitor; NNRTI = non-nucleoside reverse transcriptase inhibitor; INI = integrase inhibitor; CA-US HIV RNA = cell-associated HIV RNA.
Frequency and percent.
Median with minimum and maximum range.
“Proximal” refers to most recent CD4+ T cell count.
Summary statistics are shown for the mean of 3 baseline values as described in Elliott et. al., Lancet HIV, 2015 (19).
Disulfiram Pharmacokinetics
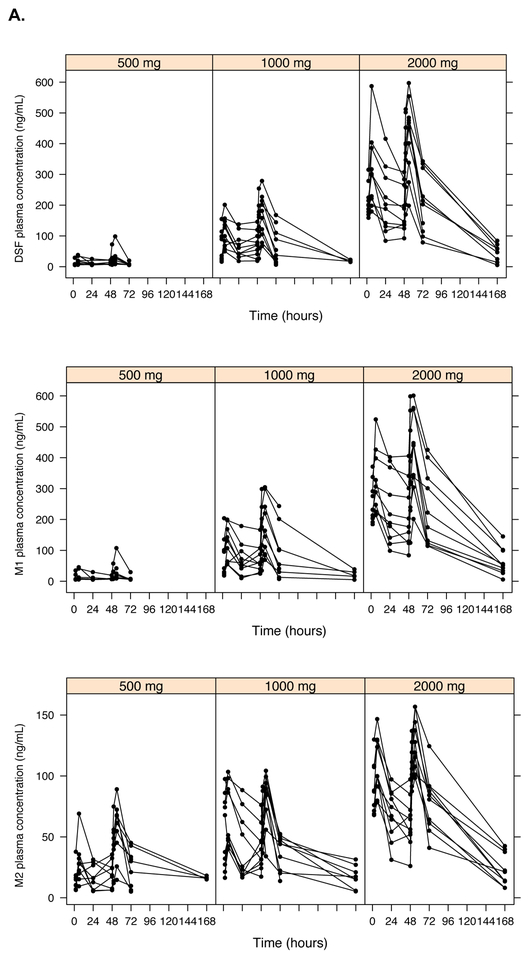
Plasma concentrations of disulfiram and four disulfiram metabolites were measured: N,N-diethyldithiocarbamate (DDTC; referred to as M1), diethyldithiocarbamate-methyl ester (DDTC-Me; M2), S-methyl-N,N-diethylthiolcarbamate sulfoxide (DETC-MeSO; M3) and carbamathione (M4). DSF and metabolite concentrations were measured on dosing day 1 (hours 0, 2, and 6), day 2 (hour 0), and day 3 (hours 0, 2, and 6), as well as on post-dosing days 4, 8, and 31 (Figure 1A). Higher concentrations of disulfiram and its metabolites were observed with increasing administered doses of disulfiram (Figure 1A). Linear PK models simultaneously incorporating all metabolites underpredicted plasma concentrations in the 2000 mg group, which initially appeared to be due to supra-proportional concentrations of disulfiram in the highest dosing group (19). However, after fitting a PK model allowing for non-linear elimination kinetics, the supra-proportional concentrations appeared to be largely explained by reduced elimination rather than increased bioavailability at the 2000 mg dose (Figure 2). Final PK model parameter estimates for DSF include a clearance of 0.53 L/hr, (CV%=36%), volume of distribution was 1.3 L and an absorption constant of 0.08 hr−1 (Table 2; Figure S1). Given the drug was completely eliminated by 72 hours, area under the curve (AUC0–72) estimates represented “cumulative” AUC estimates. The median AUC0-72 estimates using this model were 3420, 8942, and 23834 mg-hr/L for the 500, 1000, and 2000 mg DSF dosing groups, respectively (Table 3).
Figure 1.
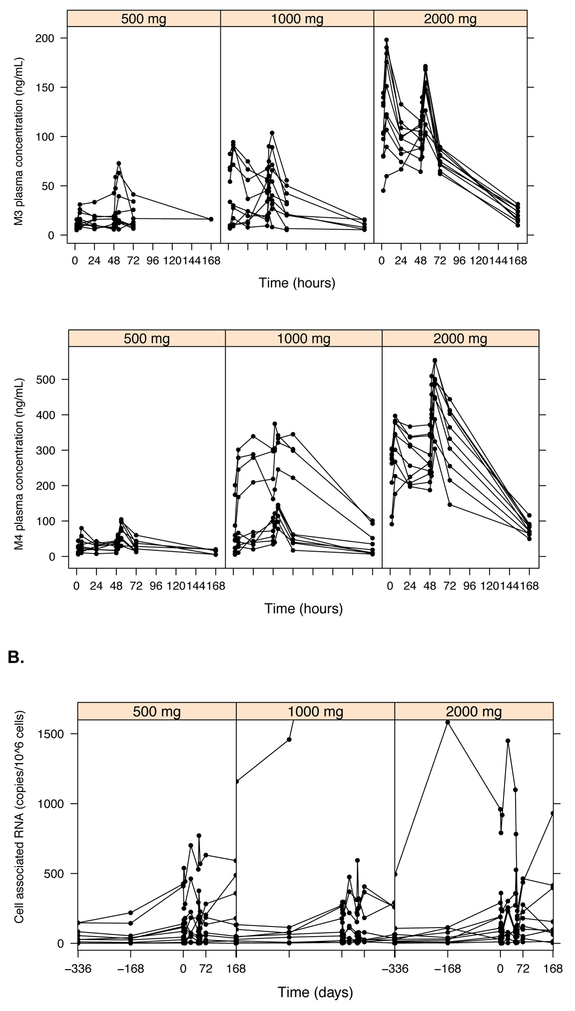
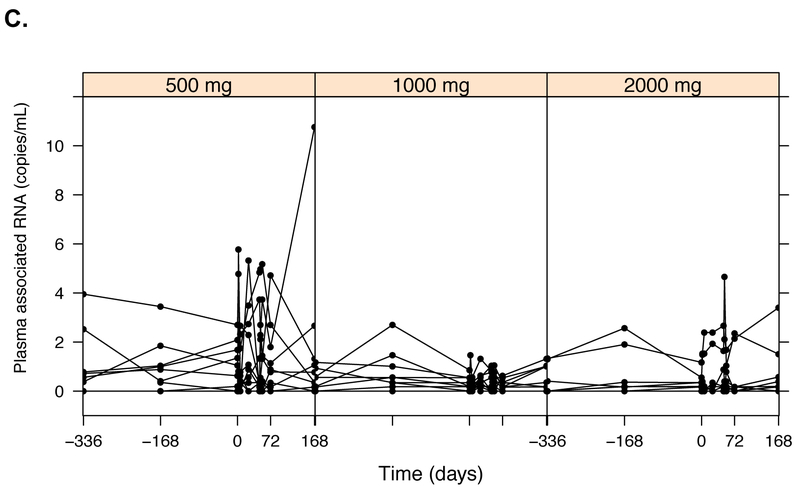
Raw pharmacokinetic and pharmacodynamic data by disulfiram dosing group. Plasma concentrations of disulfiram (DSF) and its metabolites: M1 (N,N-diethyldithiocarbamate, or DDTC), M2 (diethyldithiocarbamate-methyl ester, or DDTC-Me), M3 (S-methyl-N,N-diethylthiolcarbamate sulfoxide, or DETC-MeSO), and M4 (carbamathione) had a lower limit of detection (LLOD) of 15 ng/mL for all measured compounds (A). HIV RNA was quantified using two different assays and count-based methods (Supplemental Methods): cell-associated unspliced (CA-US) HIV RNA (copies/106 CD4+ T cells; LLOD of 1 copy/well) (B) and plasma HIV RNA (copies/mL; typical LLOD of 0.19 copies/mL) (C).
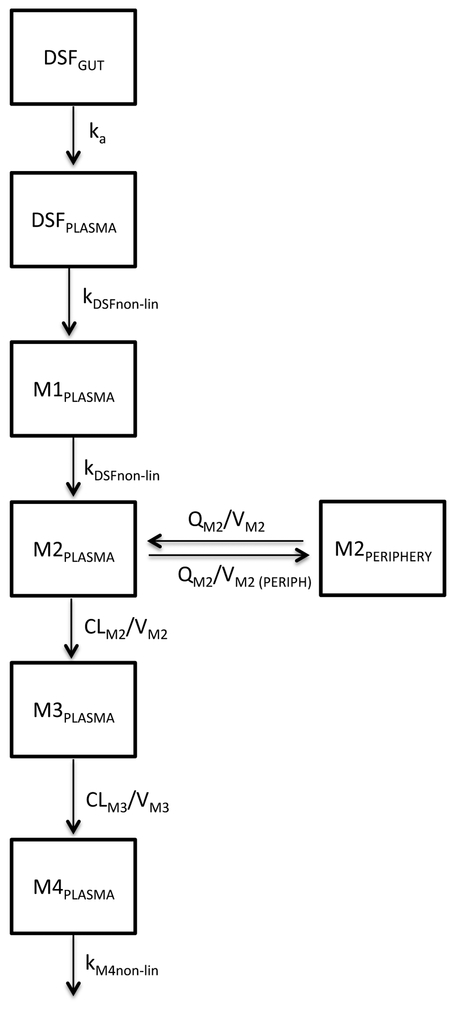
Figure 2.
Seven compartment pharmacokinetic model of disulfiram metabolism. Abbreviations: DSFGUT = amount of disulfiram in gut (mg); DSFPLASMA = amount of disulfiram in plasma (mg); M1PLASMA = amount of N,N-diethyldithiocarbamate (DDTC) in plasma (mg); M2PLASMA = amount of diethyldithiocarbamate-methyl ester (DDTC-Me) in plasma (mg); M3PLASMA = amount of S-methyl-N,N-diethylthiolcarbamate sulfoxide (DETC-MeSO) in plasma (mg); M3PERIPHERY = amount of DETC-MeSO in plasma (mg); M4PLASMA = amount of carbamathione in plasma (mg); ka = absorption constant; CL = clearance (L/hr); V = volume of distribution (L); Q = blood flow (L/hr); knon-lin = nonlinear elimination calculated using Km and Vmax terms.
| 1) |
| 2) |
| 3) |
| 4) |
| 5) |
| 6) |
| 7) |
Table 2.
Pharmacokinetic parameters measured over 72 hours (i.e., up to 24 hours after the last dose of administered disulfiram) in a model simulataneously including all metabolites.
| DSF (Disulfiram) RSE (%)a |
M1 (DDTC) RSE (%)a |
M2 (DDTC-Me) RSE (%)a |
M3 (DETC-MeSO) RSE (%)a |
M4 (Carbamathione) RSE (%)a |
|
|---|---|---|---|---|---|
| Final PK model parameters | |||||
| CL/F (L/hr)b | − | − | 0.580 (5.5) | 0.595 (6.3) | − |
| CLintrinsic (L/hr)b | 0.531 (7.7) | 0.388 (6.7) | − | − | 0.291 (4.1) |
| V (L)c | 1.29 (9.1) | 0.0906 (16.6) | 0.00404 (8.0) | 0.0647 (15.6) | 0.581 (10.3) |
| Km (ng/mL)d | 217 (9.3) | 480 (6.2) | − | − | 543 (4.3) |
| Ka (hr−1)e | 0.0752 (4.3) | − | − | − | − |
| Qf | − | − | 0.0776 (14.0) | − | − |
| Vperiph (L)g | − | − | 28.9 (17.3) | − | − |
| Additive RE (ng/mL) | 35.4 (7.1) | 41.8 (12.4) | − | 15.2 (5.3) | 82.5 (8.5) |
| Proportional RE (%) | 31.5 (9.7) | 49.5 (5.0) | 20.3 (10.1) | 40.8 (7.1) | 23.3 (19.6) |
| BV (%) in CL/F | 36 (14) | − | − | − | − |
Abbreviations: DSF = parent drug, disulfiram; M1 = metabolite 1; M2 = metabolite 2; M3 = metabolite 3; M4 = metabolite 4; DSF = disulfiram; DDTC = N,N-diethyldithiocarbamate; DDTC = diethyldithiocarbamate-methyl ester; DDTC-MeSO = S-methyl-N,N-diethylthiolcarbamate sulfoxide; PK = pharmacokinetic; CL = clearance; F = bioavailability; V = volume; RSE = residual standard error; BV = between-subject variability.
Obtained by nonparametric bootstrap with 30 samples.
Oral clearance: ; where Vmax = CLintrinsic ×Km, C = concentration of analyte, Km = is the Michaelis-Menten constant as defined below.
Volume of distribution.
Michaelis–Menten constant (concentration of drug at which elimination is half maximal).
Absorption constant.
Flow from central compartment to periphery.
Calculated peripheral volume.
Table 3.
Final PK model estimates for median AUC0–72 exposure to metabolites by dosing cohort.
| DSF (Disulfiram) RSE (%)g |
M1 (DDTC) RSE (%)g |
M2 (DDTC-Me) RSE (%)g |
M3 (DETC-MeSO) RSE (%)g |
M4 (Carbamathione) RSE (%)g |
|
|---|---|---|---|---|---|
| Final PK median AUC0–72 estimates (median and 2.5 to 97.5 percentile range) | |||||
| 500 mg group | 3,186 (3,140 – 3,281) |
2,826 (2,651 – 3,602) |
2,309 (1,563 – 3,397) |
1,563 (1,324 – 2,544) |
4,295 (4,041 – 4,923) |
| 1000 mg group | 8,386 (7,419 – 12,861) |
7,511 (4,974 – 14,886) |
4,154 (3,416 – 6,000) |
3,225 (2,102 – 5,383) |
8,640 (7,232–19,379) |
| 2000 mg group | 22,331 (18,482 – 35,102) |
20,362 (15,114 – 31,421) |
7,048 (6,248 – 9,701) |
8,643 (6,279 – 9,976) |
25,167 (16,058–32,086) |
Abbreviations: DSF = parent drug, disulfiram; M1 = metabolite 1; M2 = metabolite 2; M3 = metabolite 3; M4 = metabolite 4; DSF = disulfiram; DDTC = N,N-diethyldithiocarbamate; DDTC = diethyldithiocarbamate-methyl ester; DDTC-MeSO = S-methyl-N,N-diethylthiolcarbamate sulfoxide.
Disulfiram Pharmacokinetics/Pharmacodynamics
Cell-associated HIV RNA
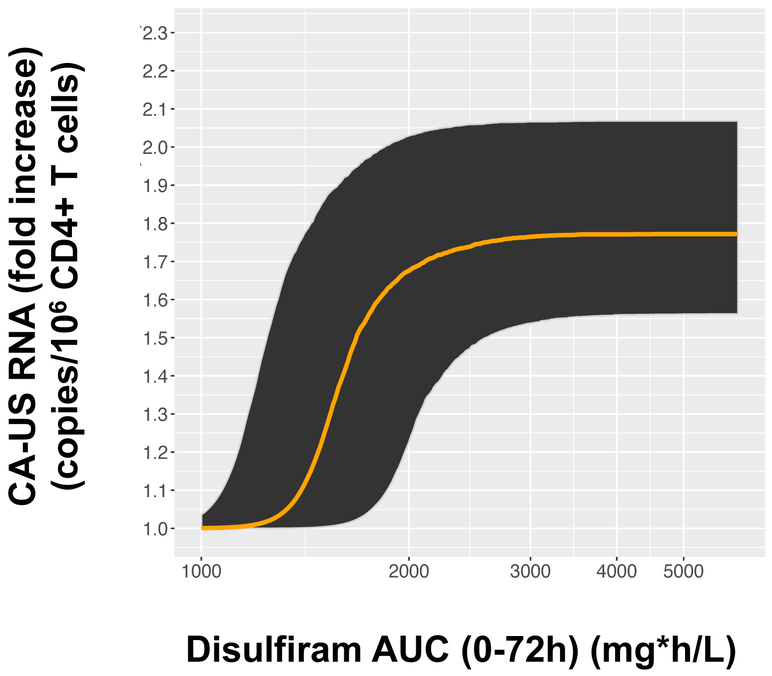
CA-US RNA was quantified at three baseline visits (B1: screening visit ~14 days prior to treatment, B2: baseline ~7 days prior to treatment, and B3: baseline visit day 0 just prior to receiving the initial dose of DSF) as well as the same timepoints as above for the PK measures (days 0–4, 8, and 31) (Figure 1B, Figure S2A). Consistent with prior latency reversal data (20), mean baseline CA-US RNA was associated with subsequent CA-US RNA at day 7 (Spearman Rho=0.88, P<0.0001) and day 31 (Spearman Rho=0.85, P<0.0001) (Figure S3). Several models were tested to estimate changes in CA-US RNA over time, including models with a parameter to account for shape (Gamma), models with Gamma but without random effects on AUC50 and Emax, and models with no covariance terms. Final PD models included Gamma (estimated to be 15.3) and three terms for covariance among the random effects for between-subject variability (Table 4, Figure S4). Models were fit for each metabolite to best predict changes in viral transcription from before to during/after disulfiram administration and account for metabolite concentrations that were below the limit of quantification (BLQ). For disulfiram, the Emax was 78%, and the AUC50 was 1600 mcg*hr/L (Table 4), and all models demonstrated a statistically significant exposure- response effect on subsequent CA-US HIV RNA levels (Table S1). In addition, there was a statistically significant exposure-response relationship on CA-US RNA; AUC0–72 DSF effect: P=6.0 × 10−17 compared a no drug effect model and P=0.013 compared to a drug effect model (Figure 3, Table S1). Statistically significant exposure-response relationships were also observed for DSF metabolites (Table S1). Models incorporating DSF exposure as measured by the AUC0–72 were statistically significant (P<0.027 for all analytes) while models estimating DSF exposure by oral admininstered dose (500, 1000, 2000 mg of DSF) were not (Table S1). We previously reported an unexplained increase in CA-US RNA at the third baseline visit (B3), which we speculated might result from changes in HIV transcription due to circadian rhythm (B1-B2 sampling occurred later in the day than B3) and/or anticipatory stress on the day of treatment (19). For these reasons, we performed post-hoc analyses including the last baseline value (B3) only rather than all three baseline values (Table S2). Models including B3 only attenuated the exposure-response effect, consistent with our previously reported results,(19).
Table 4.
Final model parametersa for the exposure-response of disulfiram on cell-associated unspliced (CA-US) HIV-1 RNA.
| Pharmacodynamic Parameters | Estimate (RSE, %a) |
|---|---|
| λbase (copies/106 CD4+ T cells) | 103 (13.6) |
| OVDP | 0.282 (6.1) |
| AUC50 (mg*h/L) | 1600 (8.9) |
| Emax (fraction increase) | 78 (13.6) |
| Gamma | 15.3 (11.5) |
| BSV (Baseline) (CV %) | 154 (16.2) |
| BSV (Overdispersion) (CV %) | 54 (24.0) |
| BSV (AUC50) (CV %) | 38 (87.7) |
| BSV (Emax) (CV %) | 47 (49.4) |
Abbreviations: λBase = mean baseline count of HIV-1 RNA, OVDP= overdispersion parameter describing the negative binomial function, AUC = area under the curve; DSF = disulfiram; Emax = concentration at which maximum efficacy observed; Gamma = parameter to account for shape; BSV = between-subject variability); CV = coefficient of variation.
Obtained by nonparametric bootstrap with 100 samples.
Figure 3.
Median fold-change exposure-response curve for cell-associated unspliced HIV-1 RNA (copies/106 CD4+ T cells) versus AUC of disulfiram concentrations (mg*h/L) over 72 hours in a model including all three baseline values. For concreteness, we plot the model’s predicted multiplicative effects as the resulting copies/106 CD4+ T cells after disulfiram administration. At each AUC value, the model estimates that 95% of the population will have a response within the shaded region, based on the estimated random effect parameters for AUC50 and Emax.
Plasma HIV RNA
Plasma HIV RNA was quantified at the same three baseline visits and PK timepoints as for CA-US HIV RNA (Figure 1C, Figure S2B). Unlike for CA-US RNA, there was no variability between the three baseline timepoints (Figure S2B). Mean baseline plasma HIV RNA levels predicted subsequent plasma HIV RNA values at day 7 (Spearman Rho=0.77, P<0.0001) and at day 31 (Spearman Rho=0.63, P=0.0002, Figure S5A). However, participants in the lowest dose 500 mg group also had the highest baseline plasma RNA levels (median 5.7 copies/mL) compared to individuals in the 1000 mg and 2000 mg groups (median 2.6 and 0.87 copies/mL, respectively, both P>0.26) (Figure S5B). The median change in plasma HIV RNA during the treatment dosing period was similar in the 500 mg group as for the 2000 mg group (-0.22 vs. −0.29 copies/mL, P=0.97), but by day 31, only the 2000 mg group demonstrated a non-statistically significant trend towards greater increases in plasma HIV RNA (1.17 vs. −0.22 copies/mL, P=0.17). Indeed, the correlation between DSF AUC0-72 and the change in plasma HIV RNA from baseline to day 31, was not statistically significant (Spearman Rho=0.31, P=0.11). Thus, the exposure-response relationship with plasma HIV RNA was less clear, and a statistically significant exposure-response was not observed between AUC0–72 and plasma HIV RNA using negative binomial regression models (Table S3).
DISCUSSION
Recent data from a Phase 2 trial demonstrated that disulfiram given at doses as high as 2000 mg daily for three days was safe and induced viral transcription in HIV+ ART-suppressed participants (19). Using population PK/PD modeling of plasma concentrations of disulfiram and four metabolites, we now find statistically significant evidence that higher cumulative exposure (AUC0-72; plasma metabolite concentrations were fully eliminated after 72 hours and thus, represent “cumulative” AUC estimates) to disulfiram and its metabolites was associated with greater increases in CA-US RNA. An exposure-response relationship was less clear for plasma HIV RNA and may have been influenced by higher mean baseline plasma HIV RNA levels in the lowest dose 500 mg group. This study is one of only a handful of HIV latency reversal trials (20–22). Though less potent than other immunomodulatory latency reversal agents, disulfiram induces HIV transcription, and this Phase 2 trial of disulfiram provides the first evidence of successful latency reversal in HIV+ individuals by a well-tolerated, well-established drug. The study also demonstrates that despite considerable inter-individual variability in the metabolism of DSF, an exposure-response effect on viral transcription can be effectively studied in HIV latency reversal trials.
Anectodotally, doses as high as 1–3 g daily of oral DSF have been given as loading doses (with prolonged administration of up to 800 mg – 1 g of DSF) in order to mitigate inter-individual variability in DSF metabolism (7) to achieve desired clinical effects (23). Disulfiram metabolism does not appear to follow first-order kinetics; prior detailed PK sampling studies have shown evidence for a second peak in plasma concentration-time profiles reflective of enterohepatic recycling (7). Our limited PK sampling here did not demonstrate a second peak per se (Figure 1A), but we used an iterative approach to account for linear and nonlinear elimination in a comprehensive seven-compartment PK model simultaneously incorporating all analytes. For the PD modeling, which required fitting separate PK models to allow for more precise estimation of individual PK parameters for each analyte (see Methods), we observed a suggestive trend between disulfiram exposure and plasma HIV RNA levels by day 31 – consistent with findings from our main Phase 2 trial publication where participants in the 2000 mg group, but not the lower doses, demonstrated a statistically significant increase in plasma HIV RNA by the end of study. Little is known about the precise molecular mechanisms by which DSF acts, but DSF has been shown to promote HIV transcription via intracellular depletion of phosphatase and tensin homolog (PTEN), upregulation of signaling through the transcription factor Akt, and the release of positive transcription elongation factor (P-TEFb), which plays an essential role in the regulation of transcription (24). It may be that disulfiram induces additional downstream pathways that continue to promote HIV transcription long after levels of disulfiram and its metabolites are no longer detectable in the blood. The discrepancy in the results for CA-US (i.e., intracellular) HIV RNA versus plasma (“cell-free”) HIV RNA could be due to differences in what each assay reflects. Specifically, that CA-US RNA represents early HIV transcription events while plasma HIV RNA reflects later steps in HIV transcription (25). Hence, since DSF acts to promote intracellular HIV transcription via increased release of P-TEFb, it may be that the exposure-response effect after short-term administration of DSF for 3 days is better captured with CA-US RNA. In contrast, plasma HIV RNA represents later steps in HIV transcription, and thus the trend seen at day 31 may reflect “late” effects of DSF on viral transcription. Finally, DSF may reverse early but not later blocks to HIV transcription in latently-infected cells (26); future cure strategies may need to include a combination of agents that act at distinct steps of HIV replication. Prospective studies of disulfiram are needed to further investigate the potential mechanisms by which disulifram acts, as well as whether the effect of disulfiram on HIV transcription is sustained. Prospective studies of disulfiram’s effects on the transcriptome, as we previously performed for the latency reversing agent vorinostat (20) are needed to define any sustained effects of disulfiram on either host or viral transcription.
We previously reported that participants receiving 2000 mg of disulfiram in the Phase 2 trial showed evidence for supra-proportional increases in disulfiram compared to participants receiving 500 mg or 1000 mg. This phenomenon was initially thought to be secondary to an increase in bioavailability at the higher dose. However, after modeling potential nonlinear elimination kinetics of downstream metabolites, these effects might be influenced by the saturation of cytochrome P450 (CYP) enzymes. Our final PK/PD models did not show a statistically significant contribution of ART regimen (e.g., protease inhibitor-, non-nuclease reverse transcriptase inhibitor-, or integrase inhibitor-based regimen) in the models, but one cannot rule out the possibility that potential drug-drug interactions with HIV medications may influence disulfiram metabolism. Indeed, a prior study evaluating disulfiram in relation to various ART regimens showed evidence for non-linear elimination kinetics even at doses as low as 62.5 mg of disulfiram daily, reflected in the study’s presented PK parameters (27). Efavirenz-containing regimens, compared to protease inhibitors, also led to lower levels of DETC-Me (S-methyl-N-N-diethylthiocarbamate), a metabolite produced between M2 and M3 as measured in our study. Thus, protease inhibitors – known CYP inducers – increased disulfiram concentrations, whereas efavirenz – a known CYP inhibitor – decreased disulfiram exposure, lending support to the hypothesis that disulfiram may follow nonlinear elimination kinetics influenced by the cytochrome P450 system.
The data presented here are based on small numbers of observations and could not clearly differentiate which metabolites were more responsible for the observed effects on HIV transcription. Though prior in vitro data demonstrated that disulfiram and M1 (DDTC), but not M2 (DDTC-Me), reactivated latent HIV (5), our PK/PD modeling of in vivo human data showed that concentrations of disulfiram and its metabolites were highly correlated, with all metabolites predicting subsequent viral transcription. Based on these data, future studies of disulfiram could potentially measure and optimize exposure to disulfiram alone (rather than all five compounds assessed here) in evaluating exposure-response to HIV transcription.
An additional limitation is that the practical importance of the exposure-response relationship we found for disulfiram and CA-US RNA is modest, because the estimated AUC50 (1600 mg*h/L) is substantially lower than the lowest AUC0–72 in our study (2108 mg*h/L). Nevertheless, because of between subject variability in AUC50, our model provides evidence that responses with a dose of 500 mg are likely to be meaningfully lower than with a dose of 1000 mg or more. Although our model estimates that over 70% of potential participants would achieve >90% of Emax at the median estimated AUC0–72 of 2270 mg*h/L in the 500 mg dose group, it also estimates that 15% of them would achieve less than a third of Emax. In contrast, at the lowest observed AUC0–72 in the 1000 mg dose group (3883 mg*h/L), 95% of potential participants would achieve >98% of Emax.
We observed a slight trend between baseline plasma HIV RNA levels and levels at day 31. These findings are consistent with our main Phase 2 trial results (19), as well as prior work from our group demonstrating that participants with higher levels of cellular HIV RNA transcription at baseline may have higher subsequent levels of HIV RNA after the administration of a histone deacetalyase (HDAC) inhibitor (20). In the study, despite significant increases in CA-US HIV RNA following the administration of an HDAC inhibitor, vorinostat, a significant change in plasma HIV RNA was not observed. This may be due to the relatively smaller dynamic range of this assay, which in chronic-treated HIV+ ART-suppressed participants can range between 0.2 – 2.6 copies/mL (28). In our study, the lowest dose (500 mg) group also had the highest median baseline plasma HIV RNA, potentially affecting the ability to demonstrate an exposure-response relationship using the negative binomial regression models. Thus, further data evaluating the effect of baseline viral transcription levels on subsequent viral transcription from future HIV latency reversal trials are needed to determine whether stratification of participants by baseline reservoir measures should be considered to balance treatment arms.
These findings provide additional support for further development of disulfiram as an important drug for future HIV cure strategies. Despite the complex nature of disulfiram metabolism, we were able to demonstrate that disulfiram produced an exposure-response effect on HIV transcription. Future HIV cure treatment will likely entail a combination therapy approach (29). The current data provide support the further investigation of disulfiram as a potential HIV cure agent to be used in combination with other more potent but toxic agents, as demonstrated in vitro (30) and is currently being explored in a follow-up trial of combination disulfiram plus vorinostat (20) (a histone deacetylase inhibitor) treatment (NCT03198559). Given its relative safety compared to other more toxic HIV latent reactivating agents and immunomodulatory agents currently being tested in clinical trials (31), and the possibility of potentially being able to administer disulfiram in conjunction with other agents to induce latent HIV transcription, may be a useful approach in future HIV eradication strategies.
METHODS
Study Participants
HIV-infected adult participants were enrolled from the SCOPE cohort at the University of California San Francisco (UCSF) and from the Department of Infectious Diseases outpatient clinic at the Alfred Hospital in Melbourne, Australia (19). Participants were enrolled in each of three sequential dosing cohorts (500, 1000, and 2000 mg) and administered oral DSF daily for three consecutive days and followed for 31 days. Inclusion criteria were confirmed HIV-1 infection, ART suppression for ≥3 years, plasma HIV-1 RNA <20 copies/mL (TaqMan version 2, Roche; NJ, USA), and CD4+T cell count >350 cells/mm3. Participants with ongoing alcohol use, significant acute illness, or concomitant use of medications containing alcohol were excluded from the study. All participants provided written informed consent, and the research was approved by the institutional review boards of the UCSF and the Alfred Human Research Ethics Committee. This study was registered with ClinicalTrials.gov (NCT01944371).
Laboratory Methods
Plasma disulfiram and metabolite concentrations were measured using ultra-performance liquid chromatography–tandem mass spectrometry (UPLC-MS/MS) (6). The four disulfiram metabolites measured were N,N-diethyldithiocarbamate (DDTC; referred to as M1), diethyldithiocarbamate-methyl ester (DDTC-Me; M2), S-methyl-N,N-diethylthiolcarbamate sulfoxide (DETC-MeSO; M3) and carbamathione (M4). The lower limit of detection was 15 ng/mL for all measured compounds. Metabolite concentrations were measured on dosing day 1 (hours 0, 2, and 6), day 2 (hour 0), and day 3 (hours 0, 2, and 6), as well as on post-dosing days 4, 8, and 31.
Cell-associated unspliced HIV RNA was measured in quadruplicate from CD4+ T cells enriched by negative selection, and HIV RNA copy numbers were standardized to cellular equivalents using an 18s RNA real time LUX polymerase chain reaction (PCR) primer set (Invitrogen) with a lower limit of detection of 1 copy equivalent per reaction, as previously described (19, 20). When there was no detectable signal, this was considered as zero copies and when there was a signal but it was <1, we assigned this a value of 0.5 copies as explained previously (19). Plasma HIV RNA was quantified using an ultrasensitive hybrid real time/digital PCR assay performed in 12 replicates from a typical plasma volume of 7 mL; this assay had a threshold sensitivity of 1 of 12 positive determinations = 0.19 copies per mL that was dependent on the actual plasma volume (32). Baseline samples were collected at the initial screening visit (B1), approximately one week prior to treatment initiation (B2), and two hours prior to the first dose of disulfiram (B3). Additional post-dosing measures were collected as above for metabolite concentrations.
Statistical Methods
Nonlinear population pharmacokinetic models incorporating plasma DSF and DSF metabolite concentrations were fit using the first-order conditional estimation with interaction (FOCEI) method using NONMEM (version 7.3; Icon Development Solutions, Dublin, Ireland). Individual parameters were assumed to be log-normally distributed, and residual variability was estimated using proportional error. Graphical diagnostics Model were used to evaluate model fit (Xpose, version 4.5.3, http://xpose.sourceforge.net) (Figures S4-S6). PK models included all metabolites simultaneously to estimate the kinetics and compartmental distribution of disulfiram metabolism. A stepwise approach was taken to model fitting. First, a structural PK model was fit to the parent compound, where DSF PK was described using one compartment disposition model with first order absorption. Linear and nonlinear elimlination mechanisms were tested, and the choice of the elimination model was supported by statistical criteria (a decrease in OFV of at least 5.99 which corresponds to a p-value of 0.01). Then each additional metabolite was incorporated into the model, one at a time, modeling elimination of the parent compound as the formation rate of the subsequent metabolite. Finally, a full model describing the plasma PK model for all five analytes for all participants was fit. Additive and proportional error models of residual variability were explored. Different residual errors were estimated for each analyte separately. The likelihood test (LRT) was used to evaluate statistical significance for inclusion of additional parameters in the nested models, assuming the objective function value (OFV) is X2 distributed. Finally, individual and population predictions of exposures at steady state were estimated in terms of the area under the concentration-time curve over 72 hours at steady state (AUC0–72) and peak concentration (Cmax) in plasma. The model was parameterized in terms of nonlinear clearance defined by Km and,Vmax, and oral volume of distribution (V). Inter-compartmental clearance (Q) and peripheral volume of distribution (V2) were tested and fitted for M2 only. Inter-individual variability (IIV) was tested on all plasma PK parameters and allowed based on the magnitude of estimates and the LRT. Given the complexity of the model, only one IIV term was added (to the parent compound). Inclusion of additional IIV terms on other structural parameters led to model instability.
Mixed effects negative binomial regression models were fit to link PK estimates with PD endpoints and account for overdispersion of low quantification values of HIV cell-associated or plasma HIV RNA levels as previously described (19). Given the above described model instability with including additional IIV terms, which would have allowed for more precise estimation of individual PK parameters for each analyte, we also fit separate PK models for each analyte alone to use in the subsequent PK/PD analyses. In order to attain additional accuracy, we included the actual BLQ values (i.e., detectable but less precise quantification for values <15 ng/mL). We did exclude however, values that were below the limit of detection (no analyte detected). Several models were evaluated; final models included cumulative drug exposure measured as the AUC over the dosing period (i.e., AUC0–72, 24 hours after the last DSF dose and when metabolite concentrations were no longer detectable). The effects of exposure were modeled as applying equally to all time points during and after disulfiram administration since fitting the dynamic concentrations as well as fitting several delay models (such as an indirect response model and an effect compartment model) to explore the time aspects of drug response did not improve the overall model fit. Maximum likelihood estimation was performed to compare minimum objective function values (OFVs) for various models. The predicted multiplicative change in CA-US RNA from baseline to during and after DSF administration for a given individual i with an AUC0–72 = Ai was calculated as:
where η3i and η4i are the random effects (individual perturbations) for AUC50 and Emax, and λtrt = the calculated mean count of HIV RNA during treatment and λbase = the calculated mean baseline count of HIV RNA. The variance of the counts was calculated using Equation 4 (Supplemental Methods), which utilizes λ and the overdispersion parameter describing the negative binomial function (OVDP). Given that in the Phase 2 trial, we observed unexplained increases in CA-US RNA at the third baseline visit (B3, morning of initial DSF dose) compared to the first two baseline vists (B1-B2, initial screening visits ~2 weeks prior to DSF), which we speculated might result from changes in HIV transcription with circadian rhythm (B1-B2 sampling occurred later in the day than B3) or caused by anticipatory stress on the day of treatment (19), we also performed post-hoc analyses using PK/PD models that included only the last baseline value.
Supplementary Material
STUDY HIGHLIGHTS
What is the current knowledge on the topic?
Despite effective antiretroviral therapy (ART), latently infected cells persist during treated HIV disease, preventing HIV eradication. Disulfiram has been shown to reactivate latently HIV-infected cells, and is being studied as a potential agent for use in HIV eradication strategies.
What question did this study address?
In this study, we investigated whether exposure to disulfiram and its metabolites predicted latent HIV reactivation in a recent Phase 2 dose-escalation study of disulfiram in HIV+ ART-suppressed participants.
What does this study add to our knowledge?
This is the first comprehensive exposure-response analysis of a clinical trial testing an agent to reactivate latent HIV for the purposes of cure. The study demonstrates the importance of careful PK monitoring to assess the effect of drug on subtle changes in viral reactivation.
How might this change clinical pharmacology or translational science?
Given its relative safety compared to other more toxic HIV latent reactivating agents and immunomodulatory agents currently being tested in clinical trials, and the possibility of potentially being able to administer disulfiram in combination with other agents to induce latent HIV transcription, our study provides support for further development of disulfiram as an important drug for future HIV cure strategies.
ACKNOWLEDGEMENTS
The authors wish to acknowledge the participation of all the study participants who contributed to this work as well as the clinical research staff of the UCSF SCOPE and Alfred Hospital research groups who made this research possible. All funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
FUNDING: This work was supported in part by the National Institutes of Health (the DARE Collaboratory [U19 AI096109], SAL: K23GM112526, RMS: KL2TR000143) the Foundation for AIDS Research (SGD: 108072–50-RGRL), and the Australian National Health and Medical Research Council (SRL: Australian National Health and Medical Research Council [NHMRC] Practitioner Fellowship; JHE: NHMRC Career Development Fellowship). This work was also supported in part with Federal funds from the National Cancer Institute, National Institutes of Health, under Contract No. HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Abbreviations:
- DSFGUT
amount of disulfiram in gut (mg)
- DSFPLASMA
amount of disulfiram in plasma (mg)
- M1PLASMA
amount of N,N-diethyldithiocarbamate (DDTC) in plasma (mg)
- M2PLASMA
amount of diethyldithiocarbamate-methyl ester (DDTC-Me) in plasma (mg)
- M3PLASMA
amount of S-methyl-N,N-diethylthiolcarbamate sulfoxide (DETC-MeSO) in plasma (mg)
- M3PERIPHERY
amount of DETC-MeSO in plasma (mg)
- M4PLASMA
amount of carbamathione in plasma (mg)
- ka
absorption constant
- CL
clearance (L/hr)
- V
volume of distribution (L)
- Q
blood flow (L/hr)
- knon-lin
nonlinear elimination calculated using Km and Vmax terms
Footnotes
Previous presentation: Preliminary data were presented in July 2017, as a poster presentation at the 9th International AIDS Society Conference on HIV Science in Paris, France, abstract #MOPEB0361.
CONFLICTS: The authors declared no competing interests for this work.
REFERENCES
- (1).Deeks SG et al. Towards an HIV cure: a global scientific strategy. Nature reviews Immunology 12, 607–14 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Chong CR, Chen X, Shi L, Liu JO & Sullivan DJ Jr. A clinical drug library screen identifies astemizole as an antimalarial agent. Nat Chem Biol 2, 415–6 (2006). [DOI] [PubMed] [Google Scholar]
- (3).Chong CR & Sullivan DJ Jr. New uses for old drugs. Nature 448, 645–6 (2007). [DOI] [PubMed] [Google Scholar]
- (4).Deeks SG HIV: Shock and kill. Nature 487, 439–40 (2012). [DOI] [PubMed] [Google Scholar]
- (5).Xing S et al. Disulfiram reactivates latent HIV-1 in a Bcl-2-transduced primary CD4+ T cell model without inducing global T cell activation. Journal of virology 85, 6060–4 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Spivak AM et al. A pilot study assessing the safety and latency-reversing activity of disulfiram in HIV-1-infected adults on antiretroviral therapy. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America 58, 883–90 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Faiman MD, Jensen JC & Lacoursiere RB Elimination kinetics of disulfiram in alcoholics after single and repeated doses. Clin Pharmacol Ther 36, 520–6 (1984). [DOI] [PubMed] [Google Scholar]
- (8).Massanella M & Richman DD Measuring the latent reservoir in vivo. J Clin Invest 126, 464–72 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Lewin SR et al. Use of real-time PCR and molecular beacons to detect virus replication in human immunodeficiency virus type 1-infected individuals on prolonged effective antiretroviral therapy. Journal of Virology 73, 6099–103 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Procopio FA et al. A Novel Assay to Measure the Magnitude of the Inducible Viral Reservoir in HIV-infected Individuals. EBioMedicine 2, 872–81 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Palmer S et al. New real-time reverse transcriptase-initiated PCR assay with single-copy sensitivity for human immunodeficiency virus type 1 RNA in plasma. J Clin Microbiol 41, 4531–6 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Rosenbloom DI, Elliott O, Hill AL, Henrich TJ, Siliciano JM & Siliciano RF Designing and Interpreting Limiting Dilution Assays: General Principles and Applications to the Latent Reservoir for Human Immunodeficiency Virus-1. Open Forum Infect Dis 2, ofv123 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Barth KS & Malcolm RJ Disulfiram: an old therapeutic with new applications. CNS Neurol Disord Drug Targets 9, 5–12 (2010). [DOI] [PubMed] [Google Scholar]
- (14).Faiman MD, Artman L & Haya K Disulfiram distribution and elimination in the rat after oral and intraperitoneal administration. Alcohol Clin Exp Res 4, 412–9 (1980). [DOI] [PubMed] [Google Scholar]
- (15).Madan A, Parkinson A & Faiman MD Identification of the human and rat P450 enzymes responsible for the sulfoxidation of S-methyl N,N-diethylthiolcarbamate (DETC-ME). The terminal step in the bioactivation of disulfiram. Drug Metab Dispos 23, 1153–62 (1995). [PubMed] [Google Scholar]
- (16).Faiman MD, Kaul S, Latif SA, Williams TD & Lunte CE S-(N, N-diethylcarbamoyl)glutathione (carbamathione), a disulfiram metabolite and its effect on nucleus accumbens and prefrontal cortex dopamine, GABA, and glutamate: a microdialysis study. Neuropharmacology 75, 95–105 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Peachey JE, Brien JF, Roach CA & Loomis CW A comparative review of the pharmacological and toxicological properties of disulfiram and calcium carbimide. J Clin Psychopharmacol 1, 21–6 (1981). [DOI] [PubMed] [Google Scholar]
- (18).Cobby J, Mayersohn M & Selliah S The rapid reduction of disulfiram in blood and plasma. J Pharmacol Exp Ther 202, 724–31 (1977). [PubMed] [Google Scholar]
- (19).Elliott JH et al. Short-term administration of disulfiram for reversal of latent HIV infection: a phase 2 dose-escalation study. Lancet HIV 2, e520–9 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Elliott JH et al. Activation of HIV transcription with short-course vorinostat in HIV-infected patients on suppressive antiretroviral therapy. PLoS pathogens 10, e1004473 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Rasmussen TA et al. Panobinostat, a histone deacetylase inhibitor, for latent-virus reactivation in HIV-infected patients on suppressive antiretroviral therapy: a phase 1/2, single group, clinical trial. Lancet HIV 1, e13–21 (2014). [DOI] [PubMed] [Google Scholar]
- (22).Vibholm L et al. Short-Course Toll-Like Receptor 9 Agonist Treatment Impacts Innate Immunity and Plasma Viremia in Individuals With Human Immunodeficiency Virus Infection. Clin Infect Dis 64, 1686–95 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Talbott GD & Gander O Antabuse, 1973. Md State Med J 22, 60–3 (1973). [PubMed] [Google Scholar]
- (24).Doyon G, Zerbato J, Mellors JW & Sluis-Cremer N Disulfiram reactivates latent HIV-1 expression through depletion of the phosphatase and tensin homolog. AIDS 27, F7–F11 (2013). [DOI] [PubMed] [Google Scholar]
- (25).Pasternak AO, Lukashov VV & Berkhout B Cell-associated HIV RNA: a dynamic biomarker of viral persistence. Retrovirology 10, 41 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Yukl SA et al. HIV latency in isolated patient CD4(+) T cells may be due to blocks in HIV transcriptional elongation, completion, and splicing. Sci Transl Med 10, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).McCance-Katz EF et al. Interaction of disulfiram with antiretroviral medications: efavirenz increases while atazanavir decreases disulfiram effect on enzymes of alcohol metabolism. Am J Addict 23, 137–44 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Eriksson S et al. Comparative analysis of measures of viral reservoirs in HIV-1 eradication studies. PLoS Pathog 9, e1003174 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Katlama C et al. Barriers to a cure for HIV: new ways to target and eradicate HIV-1 reservoirs. Lancet 381, 2109–17 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Laird GM et al. Ex vivo analysis identifies effective HIV-1 latency-reversing drug combinations. J Clin Invest 125, 1901–12 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Delagreverie HM, Delaugerre C, Lewin SR, Deeks SG & Li JZ Ongoing Clinical Trials of Human Immunodeficiency Virus Latency-Reversing and Immunomodulatory Agents. Open Forum Infect Dis 3, ofw189 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Somsouk M et al. The immunologic effects of mesalamine in treated HIV-infected individuals with incomplete CD4+ T cell recovery: a randomized crossover trial. PLoS One 9, e116306 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.