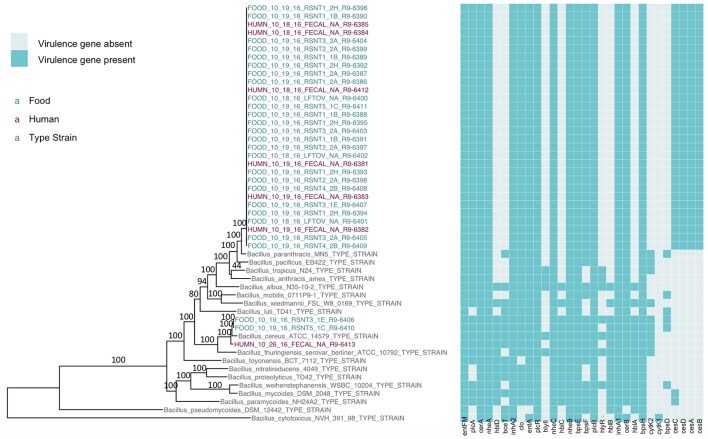
Figure 1.
Maximum likelihood phylogeny of core SNPs identified in 33 isolates sequenced in conjunction with a B. cereus outbreak, as well as genomes of the 18 currently recognized B. cereus group species (shown in gray). Core SNPs were identified in all genomes using kSNP3. Heatmap corresponds to presence/absence of B. cereus group virulence genes detected in each sequence using BTyper. Tip labels in maroon and teal correspond to the seven human clinical isolates and 26 isolates from food sequenced in conjunction with this outbreak, respectively. Phylogeny is rooted at the midpoint, and branch labels correspond to bootstrap support percentages out of 500 replicates. Due to the short lengths and low bootstrap support (all values <10) of branches within the outbreak clade, bootstrap support percentages are not shown on branches within the outbreak clade.