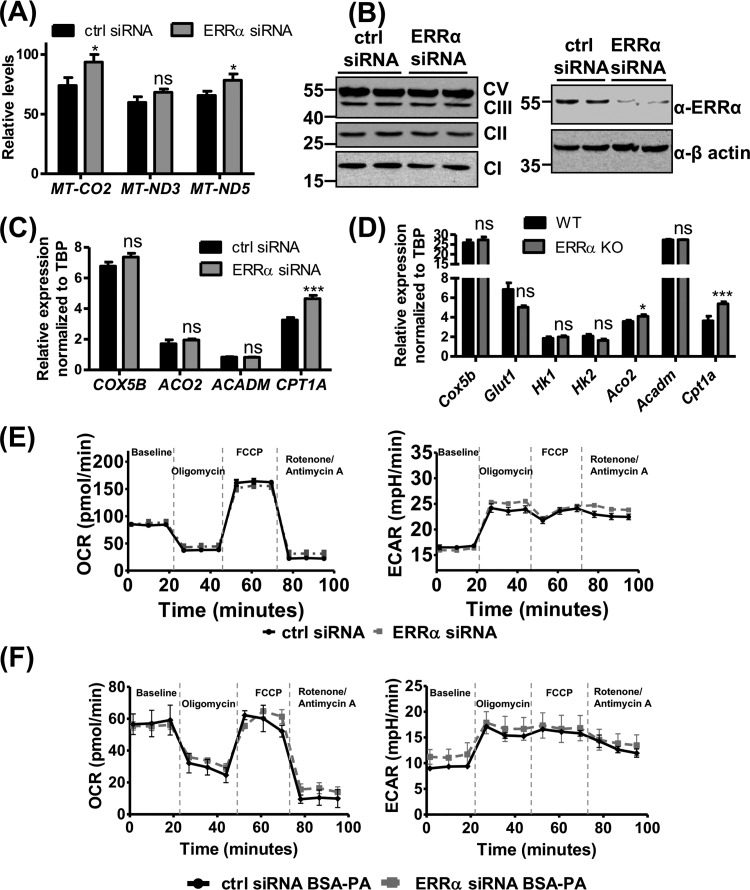
FIG 5.
ERRα does not affect endothelial metabolism under normal growth conditions. (A) Mitochondrial DNA content in control and ERRα knockdown ECs (n = 3). (B) Protein expression levels of ERRα and mitochondrial oxidative phosphorylation complexes in control and ERRα knockdown HUVEC (n = 2). (C and D) Gene expression of known ERRα metabolic target genes in ERRα-KO versus WT murine lung ECs (C) and ERRα knockdown versus control HUVEC (D) (n = 6). (E) Representative graphs of mitochondrial respiration, with glucose-pyruvate as the energy source measured as the oxygen consumption rate (OCR) (left) and glycolysis measured as the extracellular acidification rate (ECAR) (right) in ERRα knockdown versus control HUVEC at baseline and upon sequential drug treatment. Unpaired Student's t test was used (n = 3, each performed in 5 or 6 technical replicates). (F) Representative graphs of mitochondrial respiration with palmitate (BSA-PA) as the energy source measured as the oxygen consumption rate (OCR) (left) and glycolysis measured as the extracellular acidification rate (ECAR) (right) in ERRα knockdown versus control HUVEC at baseline and upon sequential drug treatment (n = 3). *, P < 0.05; ***, P < 0.001, unpaired Student's t test. ns, not significant.