Abstract
CRISPR/Cas9 is a powerful genome editing system but uncontrolled Cas9 nuclease expression triggers off-target effects and even in vivo immune responses. Inspired by synthetic biology, here we built a synthetic switch that self-regulates Cas9 expression not only in the transcription step by guide RNA-aided self-cleavage of cas9 gene, but also in the translation step by L7Ae:K-turn repression system. We showed that the synthetic switch enabled simultaneous transcriptional and translational repression, hence stringently attenuating the Cas9 expression. The restricted Cas9 expression induced high efficiency on-target indel mutation while minimizing the off-target effects. Furthermore, we unveiled the correlation between Cas9 expression kinetics and on-target/off-target mutagenesis. The synthetic switch conferred detectable Cas9 expression and concomitant high frequency on-target mutagenesis at as early as 6 h, and restricted the Cas9 expression and off-target effects to minimal levels through 72 h. The synthetic switch is compact enough to be incorporated into viral vectors for self-regulation of Cas9 expression, thereby providing a novel ‘hit and run’ strategy for in vivo genome editing.
INTRODUCTION
CRISPR/Cas9 is a powerful genome editing system comprising the Cas9 nuclease and chimeric single guide RNA (gRNA) (1). Guided by the spacer sequence on gRNA that can be designed in a customizable fashion, the Cas9/gRNA complex recognizes the chromosomal protospacer-adjacent motif (PAM) and binds to proximal complementary sequence (protospacer), thereby triggering a double strand break (DSB) at the target sequence (1–3). Such CRISPR/Cas9-guided DSB promotes foreign gene integration near the DSB site via homology directed repair (HDR), or results in small nucleotide insertions/deletions (indels) via non-homologous end joining (NHEJ). Such property renders CRISPR/Cas9 a versatile tool for programmable genome engineering of eucaryotic and procaryotic cells (4–8) as well as for cell and gene therapy (9,10).
Early studies have mainly utilized plasmids to express Cas9 derived from Streptococcus pyogenes (referred to as Cas9 thereafter) and gRNA within the cells to induce DSB at the predetermined genomic site (1–3). However, in this format the overexpressed Cas9 persists for an extended period of time and triggers indel mutation at off-target sites (11,12). Alternatively, Cas9 can be delivered in the form of protein (13) or mRNA (14,15), together with the chemically synthesized gRNA, to reduce the off-target effects. The Cas9 protein/gRNA ribonucleoprotein (RNP) approach is gaining popularity for in vitro gene editing (16–18) and recent studies have demonstrated its applications in in vivo genome engineering (19,20). However, efficient in vivo delivery of Cas9/gRNA RNP remains problematic due to challenges in delivering macromolecular complex into nucleus in vivo (21–23).
Conversely, a wide variety of strategies have been developed to control the expression level and duration of functional Cas9 (for review see (8,24,25)). Among these approaches, a self-restrict lentivirus (LV) vector was developed which, in addition to the specific targeting gRNA, co-expresses an extra gRNA that targets the cas9 gene, hence triggering cas9 self-cleavage and transcription abortion (26,27). Using the similar strategy, Ruan et al. developed a self-destructing CRISPR/Cas9 system that cleaves cas9 gene by a pair of gRNA and limits the cas9 transcription (28).
Synthetic biology is a burgeoning discipline that aims to engineer biological devices such as synthetic switch to operate gene expression control in the transcription, translation and post-translation levels (29,30). One of the synthetic devices exploits the L7Ae:K-turn system in which the RNA-binding protein L7Ae binds tightly to RNA motif known as kink-turn (K-turn) and represses subsequent translation (31). The L7Ae:K-turn interaction was first harnessed to construct an OFF switch to repress mRNA translation by inserting K-turn motifs at the 5′ UTR of a reporter gene (31) and was later leveraged to assemble synthetic switch (32–34), multi-input logic gate (35) and complex synthetic circuits (36). However, the L7Ae:K-turn system has yet to be explored for Cas9 expression control.
Here, we hypothesized that simultaneous inhibition of transcription and translation can confer more stringent control of Cas9 expression and minimize off-target effects. Therefore, we built a synthetic switch that combines (i) the Cas9/gRNA to target and self-destruct the cas9 gene for transcription abortion and (ii) the L7Ae:K-turn system to self-repress the cas9 mRNA translation. We showed that the synthetic switch enabled simultaneous transcriptional and translational repression, hence synergistically and stringently restricting the Cas9 expression. The attenuated Cas9 expression induced high efficiency on-target indel mutation while minimizing the off-target effects. Furthermore, we unveiled that the synthetic switch-regulated expression triggered high frequency on-target mutation at as early as 6 h post-transfection while restricting the Cas9 expression and off-target mutation to minimal levels through 72 h. The synthetic switch is compact enough to be incorporated into viral vectors, hence holding great promise for self-regulation of Cas9 expression and in vivo genome editing.
MATERIALS AND METHODS
Plasmids construction
The pK-CLY and pCLY plasmids were constructed using pUseAmp(+) (Merck Millipore) that harbored the CMV promoter/enhancer, multiple cloning site and bGH poly A signal as the backbone. We PCR-amplified the human codon-optimized Cas9 with the N-terminal SV40 nuclear localization signal (NLS) from pX330 (Addgene (2)) and amplified 2 repeats of K-turn motifs from pTK095 (Addgene (36)). Using overlap PCR, we amplified the DNA fragment encoding L7Ae, porcine teschovirus-1 2A (P2A) ribosome skipping peptide, enhanced yellow fluorescent protein (EYFP) from pTK095 and simultaneously added an additional 5′ P2A sequence by primer design. To yield pK-CLY, we subcloned these DNA fragments into pUseAmp(+) by enzyme digestion/ligation in the following order: K-turn motifs, NLS-Cas9 and P2A-L7Ae-P2A-EYFP. To yield pCLY, we performed similar cloning steps except that K-turn motifs were not included.
In parallel, the sequence encoding the human U6 promoter, gRNA scaffold and terminator was PCR-amplified from pX335 (Addgene (2)) and subcloned into TA vector (Invitrogen) to yield pTA-U6gRNA-Term vector. The 20 bp spacer DNA duplexes targeting the human VEGFA1 and cas9 gene, or targeting no sequence (ϕ) were synthesized, cloned into the gRNA scaffold by enzyme digestion and sequenced to confirm the correct cloning. The spacer targeting VEGFA1 was identical to that reported previously (37). The resultant gRNA expression vectors were designated pTA-gVgC, pTA-gV and pTA-gϕ, respectively.
Cell culture and transfection
HEK293, HeLa and HepG2 cells were routinely cultured in high-glucose DMEM medium supplemented with 10% fetal bovine serum (FBS) (Invitrogen). The cells were seeded to 6-well plates (5 × 105 cells/well for HEK293, and 3 × 105 cells/well for HeLa and HepG2) and cultured for 24 h. We then co-transfected pCLY or pK-CLY (4 μg/well) with one of the 3 gRNA expression plasmids (2 μg/well) into cells using lipofectamine 3000 (Thermo Fisher Scientific) and analyzed indel mutation frequencies, EYFP and Cas9 expression at 24 h post-transfection. To analyze the kinetics of Cas9 expression and on-target/off-target mutagenesis, the cells were harvested at different time points. For analyses at 48 and 72 h, the cells were replenished with fresh medium at 24 h post-transfection and continued to be cultured.
Fluorescence microscopy and flow cytometry
At 24 h post-transfection, cells were observed under the fluorescence microscope (ECLIPSE Ti2-E, Nikon) for yellow fluorescence emission. The percentage of cells emitting fluorescence and mean fluorescence intensity (FI) of each sample were measured 3 times by counting 10,000 cells in each measurement. Multiplying the mean FI by the number of EYFP+ cells yielded the total FI (expressed in arbitrary units). All the total FI data were normalized to that in the group transfected with pCLY and pTA-gϕ (CLY/gϕ group) to yield the relative EYFP intensity (%).
Western blot analysis of Cas9 protein
Proteins were extracted from HEK293 cells using the RIPA buffer, separated by 10% SDS-PAGE under denature conditions, electrotransferred to nitrocellulose membranes and were subjected to Western blot following standard procedures. The primary antibody was GTX53807 mAb (1:3000 dilution, GeneTex) to probe Cas9 or GTX109639 pAb (1:5000 dilution, GeneTex) to detect the internal control β-actin. The secondary antibodies were ab6789 (1:10 000 dilution, Abcam) for Cas9 and ab6721 (1:10 000 dilution, Abcam) for β-actin. To ensure the consistency and accuracy of quantification, for each experiment all electrotransferred nitrocellulose membranes containing different samples were incubated with the antibodies in the same container at the same time, developed at the same time using the Western Lightning Plus-ECL enhanced chemiluminescence substrate (Perkin Elmer) and imaged using GeneGnome HR (Syngene). The band intensities were scanned, analyzed by ImageJ and normalized to those of the internal control β-actin. All the band intensity data were normalized again to that of the control group transfected with pCLY and pTA-gϕ (CLY/gϕ group).
Analysis of indel mutation frequency
Since gRNAVEGFA1 was designed as described (37), we selected the on-target site and off-target sites (OT-1, OT-2 and OT-3) that were reported to have the highest DSB scores (37) for indel mutation analysis. The frequency of indel mutations was analyzed by mismatch cleavage assay using the GeneArt® Genomic Cleavage Detection Kit (Thermo Fisher Scientific) following the manufacturer's instructions. Briefly, the genomic DNA was extracted by Cell Lysis buffer (Thermo Fisher Scientific) and the loci where the DSB occurred were amplified by PCR using the AmpliTaq Gold 360 Master Mix (Thermo Fisher Scientific) with specific primers (Supplementary Table S1). The PCR amplicons were denatured and re-annealed, followed by digestion with the Detection Enzyme (Thermo Fisher Scientific). The resultant DNA fragments were analyzed by 2% TBE agarose gel electrophoresis and stained by ethidium bromide. The band intensities were determined by scanning densitometry using ImageJ and the frequencies of indel mutation were calculated following the manufacturer's instructions. All the indel mutation frequency data were subtracted from that of the control group (the cells co-transfected with pCLY and pTA-gϕ).
Statistical analysis
All data obtained from experiments performed at 24 h post-transfection are averages±standard deviation (SD) of 3 independent culture experiments. The data from time-course experiments are representative of two independent culture experiments. Statistical comparisons were performed using the two-tailed Student's t-test. P < 0.05 was considered statistically significant.
RESULTS
Design of synthetic switch to restrict Cas9 transcription and translation
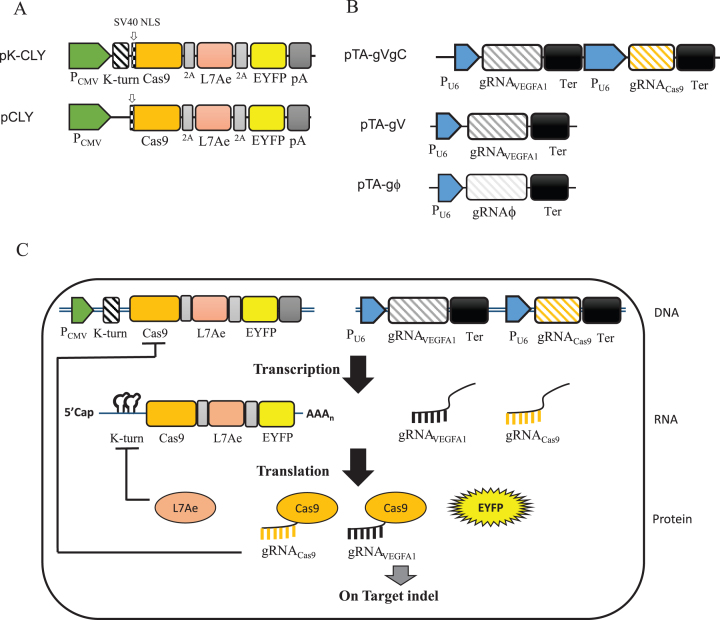
To build a synthetic switch that combines Cas9/gRNA self-destructing the cas9 gene and L7Ae:K-turn system to control Cas9 expression in the transcription and translation level, we first constructed two plasmids, pK-CLY and pCLY, which co-expressed Cas9, L7Ae and EYFP that were linked in tandem by the P2A sequence, with (pK-CLY) or without (pCLY) two repeats of K-turn motifs at the 5′ end (Figure 1A). In previous pioneering studies exploring/developing Cas9, VEGFA1 was commonly investigated as the target genes (2,37–40), thus we first chose VEGFA1 as the target. We constructed pTA-gVgC (Figure 1B) that co-expressed two different gRNAs: gRNAVEGFA1 targeting human VEGFA1 gene and gRNACas9 targeting the plasmid-borne cas9 gene (Supplementary Figure S1A). The gRNAVEGFA1 sequence was identical to that described previously (37) so that the on-target and off-target mutation sites could be selected as described earlier (37). As controls, we also constructed pTA-gV expressing gRNAVEGFA1 and pTA-gϕ expressing a scramble gRNA (gRNAϕ) that targets no sequence on the chromosomes or plasmids (Figure 1B).
Figure 1.
Design of the synthetic switch to restrict Cas9 transcription and translation. (A) Plasmids to co-express Cas9, L7Ae and EYFP, with (pK-CLY) or without (pCLY) two repeats of K-turn at the 5′ end. The three genes were linked in tandem by P2A sequences and driven by CMV promoter/enhancer. Cas9 was fused with an SV40 nuclear localization signal (NLS). pA, poly A signal. (B) Plasmids to express the gRNA. pTA-gVgC co-expressed two different gRNAs: gRNAVEGFA1 targeting human VEGFA1 gene on human chromosome 6 and gRNACas9 targeting the plasmid-borne Cas9 gene. pTA-gV expressed gRNAVEGFA1 while pTA-gϕ expressed a scramble gRNA (gRNAϕ) that targets no sequence on the chromosomes or plasmids. All gRNAs were driven by human U6 promoter. Ter, transcription termination sequence. (C) Schematic illustration of how the synthetic switch self-regulates Cas9 expression in the transcription and translation steps. Co-transfection of pK-CLY and pTA-gVgC into cells would lead to co-transcription of Cas9, L7Ae, EYFP, gRNAVEGFA1 and gRNACas9. Cas9 is translated and orchestrates with gRNAVEGFA1 to induce on-target indel mutation at VEGFA1 site while also associating with gRNACas9 to inhibit Cas9 transcription by cleaving the plasmid-borne Cas9 gene. Additionally, L7Ae is translated and in turn binds the K-turn motifs at the 5′ UTR of mRNA to suppress mRNA translation.
We envisioned that co-transfection of pK-CLY and pTA-gVgC into cells would lead to co-transcription of Cas9, L7Ae, EYFP, gRNAVEGFA1 and gRNACas9 (Figure 1C). Cas9 is translated and orchestrates with gRNAVEGFA1 to induce on-target indel mutation at VEGFA1 site while also associating with gRNACas9 to abort Cas9 transcription by cleaving the plasmid-borne cas9 gene. Additionally, L7Ae is translated and in turn binds the K-turn motifs at the 5′ UTR of mRNA to self-suppress mRNA translation (Figure 1C). EYFP is translated as a reporter. This synthetic switch would confer concurrent transcriptional and translational repression of Cas9 and EYFP. In contrast, co-transfection of pCLY and pTA-gVgC would restrict Cas9 transcription only by cleaving the cas9 gene but would not prevent cas9 mRNA translation due to the lack of K-turn motifs.
Verification of restricted Cas9 expression
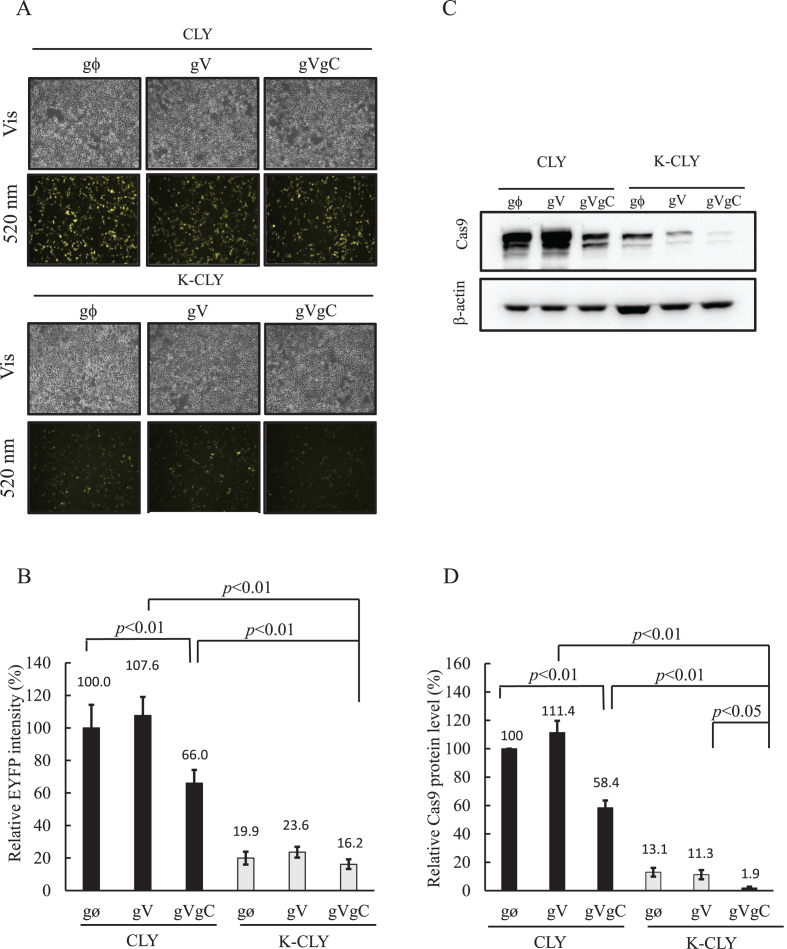
To verify the design, we first co-transfected pCLY with pTA-gϕ (CLY/gϕ group), pTA-gV (CLY/gV group) or pTA-gVgC (CLY/gVgC group) into HEK293 cells. Fluorescence micrographs illustrated evident EYFP expression at 24 h post-transfection in all three groups, indicating efficient transfection (upper panel, Figure 2A). However, flow cytometry analysis (Figure 2B) revealed that the EYFP intensity in the CLY/gVgC group was only 66.0 ± 8.1% that of the CLY/gϕ group (defined as 100%), attesting that Cas9 and gRNACas9 coordinated to cleave the plasmid-borne cas9 gene and hence mitigated downstream EYFP expression. When we co-transfected pK-CLY with pTA-gϕ (K-CLY/gϕ group), pTA-gV (K-CLY/gV group) or pTA-gVgC (K-CLY/gVgC group), the EYFP intensity was further reduced to ≈16–24% that of the CLY/gϕ group (Figure 2A and B), proving that the K-turn motifs on the pK-CLY-transcribed mRNA enabled L7Ae binding and spontaneous translational repression of EYFP.
Figure 2.
Verification of synthetic switch design. (A) Microscopic observation of cells. (B) Relative EYFP intensity. (C) Western blot analysis of Cas9 expression. (D) Relative Cas9 expression. We first co-transfected pCLY with pTA-gϕ (CLY/gϕ group), pTA-gV (CLY/gV group) or pTA-gVgC (CLY/gVgC group) into HEK293 cells. In parallel, we co-transfected pK-CLY with pTA-gϕ (K-CLY/gϕ group), pTA-gV (K-CLY/gV group) or pTA-gVgC (K-CLY/gVgC group) into HEK 293 cells. At 24 h post-transfection, cells were observed with a fluorescence microscope (magnification, 100×) and analyzed by flow cytometry. All total fluorescence intensity data were normalized to that in the CLY/gϕ group to yield the relative EYFP intensity (%). The proteins in each group were analyzed by Western bot at the same time under same conditions. All band intensities were scanned, analyzed by ImageJ, normalized to that of the internal control β-actin and normalized again to that of the CLY/gϕ group. All data are representative of three independent culture experiments.
Whether the synthetic switch concomitantly inhibited Cas9 expression was confirmed by western blot at 24 h post-transfection. Figure 2C shows robust Cas9 expression in the groups without gRNA-mediated self-cleavage and L7Ae:K-turn repression (CLY/gϕ and CLY/gV groups). We performed scanning densitometry and defined the Cas9 expression in the CLY/gϕ group as 100% (Figure 2D). With the gRNA targeting cas9 gene, the CLY/gVgC group expressed 58.4 ± 1.5% Cas9. By L7Ae:K-turn repression only, Cas9 expression in the K-CLY/gϕ and K-CLY/gV groups dropped to 13.1 ± 3.2% and 11.3 ± 3.2%, respectively. With the synthetic switch combining gRNA-mediated self-cleavage and L7Ae:K-turn repression, Cas9 expression in the K-CLY/gVgC group precipitously dropped to 1.9 ± 0.8% (Figure 2C and D). Figure 2 collectively confirms that L7Ae:K-turn alone suppressed EYFP and Cas9 translation, and acted in concert with the gRNA-mediated self-cleavage to substantially attenuate the expression.
Attenuated Cas9 expression induced on-target mutation while reducing off-target effects
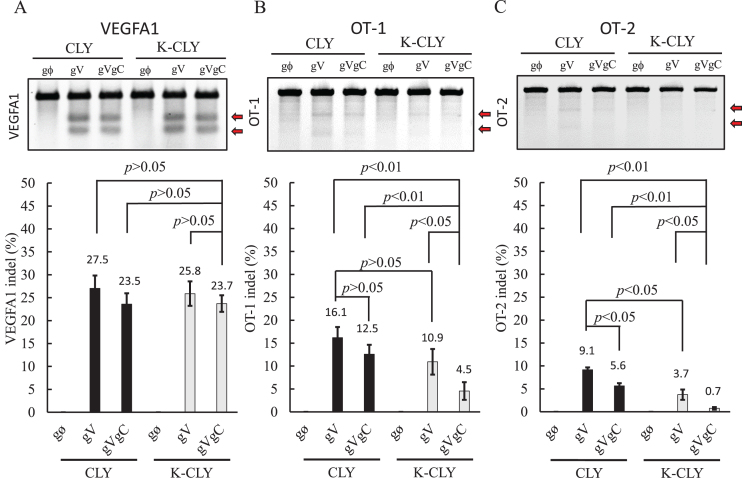
To examine whether the attenuated Cas9 expression sufficed to induce on-target indel mutation and minimize off-target effects, we co-transfected pCLY or pK-CLY with one of the three gRNA expression plasmids into HEK293 cells as in Figure 2 and analyzed the indel frequencies at the target VEGFA1 site by mismatch cleavage assay. For off-target effects, we analyzed two corresponding off-target sites OT-1 and OT-2 (Supplementary Figure S1B) that were reported to have the highest DSB scores (37) (DSB score positively correlates with indel formation (40)). The indel mutation assay (Figure 3A) revealed that the on-target indel frequency in the K-CLY/gVgC group (23.7 ± 1.8%) was statistically similar (P > 0.05) to those in the CLY/gV (27.5 ± 2.7%), CLY/gVgC (23.5 ± 2.3%) and K-CLY/gV (25.8 ± 2.6%) groups.
Figure 3.
On-target/off-target mutagenesis in HEK293 cells. (A) Indel frequencies at VEGFA1 site. (B) Indel frequencies at off-target 1 (OT-1) site on chromosome 5. (C) Indel frequencies at OT-2 site on chromosome 5. We repeated experiments as in Figure 2 and extracted the genomic DNA for indel mutation analysis using the GeneArt® Genomic Cleavage Detection Kit. The cleavage was visualized by agarose gel electrophoresis (upper panels) and the indel frequencies (lower panels) were analyzed as described in Methods. The data represent the averages±SD of three independent culture experiments.
At the OT-1 (Figure 3B) and OT-2 (Figure 3C) sites, the robust Cas9 expression in the CLY/gV group induced high indel frequencies of 16.1 ± 2.3% and 9.1 ± 0.5%, respectively. Self-destructing cas9 gene by the CLY/gVgC group to some extent reduced indel frequencies at OT-1 (12.5 ± 2.0%) and OT-2 (5.6 ± 0.5%). With L7Ae:K-turn repression alone, the K-CLY/gV group also reduced the indel frequencies to 10.9 ± 2.8% at OT-1 and 3.7 ± 1.1% at OT-2. With the synthetic switch, the K-CLY/gVgC group significantly (P < 0.01) reduced the indel frequencies to 4.5 ± 1.9% at OT-1 and 0.7 ± 0.2% at OT-2. Also, we assessed the indel frequencies at OT-3 that had the third highest DSB score (37) and found that the K-CLY/gVgC group triggered negligible indel mutation at OT-3 (Supplementary Figure S2).
Besides VEGFA1, EMX1, RUNX1 and FNACF are commonly investigated for on-target/off-target mutagenesis (2,37–40), thus we also investigated the on-target/off-target indel mutagenesis at these genes (Supplementary Figure S3). Moreover, the sensitivity of the mismatch cleavage assay we used was considered relatively low (41,42), therefore we employed an additional method (TIDE, tracking of indel by decomposition (43)) to analyze the on-target/off-target indel mutagenesis at different sites of VEGFA1, EMX1, RUNX1 and FANCF genes (Supplementary Figure S4). Supplementary Figures S3 and S4 further confirm that the synthetic switch induced similar on-target indel frequencies and lower off-target indel frequencies when compared with controls. These data altogether attest that attenuating Cas9 expression in the transcription and translation steps minimized off-target effects without sacrificing the on-target mutagenesis.
Cas9 restriction strategy can be adapted to different cells
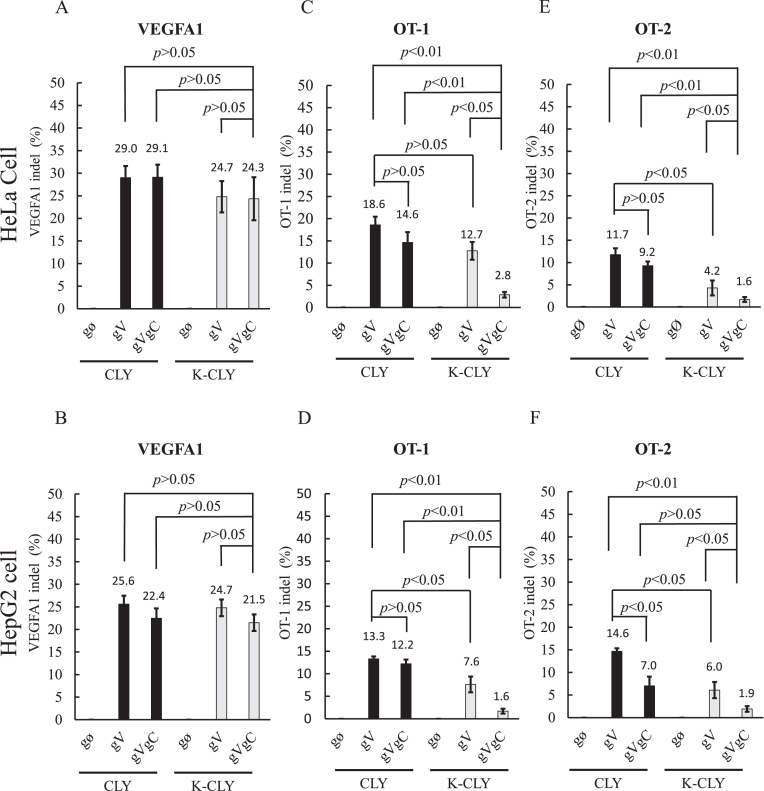
To assess whether this approach could be extrapolated to other human cells, we repeated the experiments by co-transfecting the plasmids into HeLa or HepG2 cells. In agreement with the data in HEK293 cells, the synthetic switch in the K-CLY/gVgC group effectively repressed the EYFP expression in both HeLa and HepG2 cells (Supplementary Figure S5). Concurrently, the K-CLY/gVgC group provoked statistically similar on-target indel frequencies at the VEGFA1 site (Figure 4A and B) and elicited lower off-target effects at the OT-1 and OT-2 sites (Figure 4C–F) when compared with CLY/gV, CLY/gVgC and K-CLY/gV groups.
Figure 4.
On-target/off-target mutagenesis in HeLa and HepG2 cells. (A, B) On-target indel frequencies at VEGFA1 site in HeLa and HepG2 cells. (C, D) Off-target indel frequencies at OT-1 site in HeLa and HepG2 cells. (E, F) Off-target indel frequencies at OT-2 site in HeLa and HepG2 cells. We transfected HeLa and HepG2 cells as in Figure 2 and analyzed the indel frequencies as in Figure 3. The data represent the averages ± SD of three independent culture experiments.
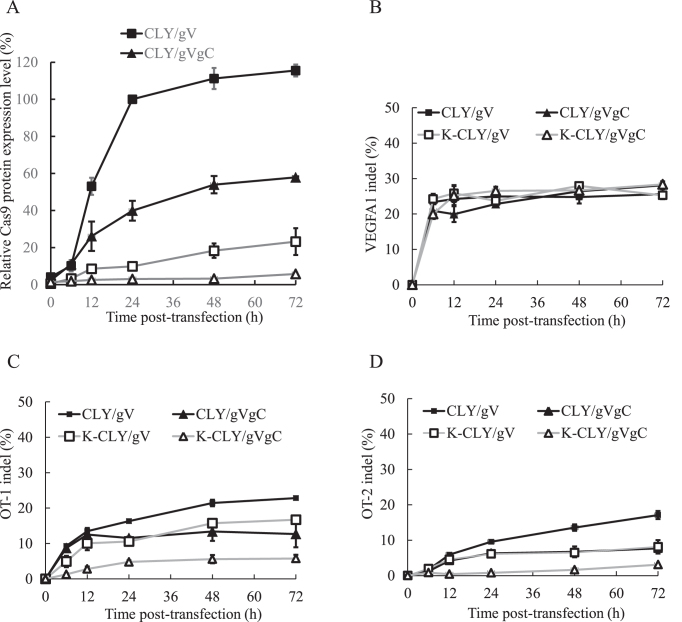
Kinetics of Cas9 protein expression and on-target/off-target mutagenesis
The aforementioned experiments demonstrated that the synthetic switch effectively reduced Cas9 expression while triggering on-target mutation. It is thus of interest to examine the kinetics of Cas9 expression and its correlation with on-target/off-target mutagenesis. To this end, we repeated the experiments in Figure 2 by co-transfecting plasmids into HEK293 cells and analyzed the Cas9 expression and indel frequencies as in Figures 2 and 3 at different time points.
By defining the Western blot band intensities of CLY/gV group at 24 h as 100%, we found detectable Cas9 expression in the CLY/gV group at as early as 6 h (10.3 ± 1.0%), which increased to 115.5 ± 3.1% at 72 h (Figure 5A). By self-cleaving the cas9 gene, the Cas9 expression in the CLY/gVgC group was also detectable at 6 h (11.0 ± 0.9%) and increased to 57.8 ± 0.5% at 72 h, suggesting that self-cleaving the cas9 gene was unable to completely abort the expression. This was likely because the cleavage needs the expression of functional Cas9, at which time some residual cas9 mRNA had been transcribed. The residual Cas9 mRNA continued to be translated, leading to the rise of Cas9 levels. With the L7Ae:K-turn translational repression, the Cas9 level in the K-CLY/gV group was 3.2 ± 0.9% at 6 h and gradually increased to 9.8 ± 1.0% at 24 h and 23.1 ± 7.2% at 72 h, suggesting that the L7Ae:K-turn system still allowed for leaky translation of the mRNA. By combining the self-cleavage and L7Ae:K-turn repression, the synthetic switch-regulated Cas9 expression in the K-CLY/gVgC group was attenuated to 1.8±0.7% at 6 h and remained as low as 3.0 ± 0.3% at 24 h and 5.7 ± 0.6% at 72 h (Figure 5A).
Figure 5.
Kinetics of Cas9 protein and on-target/off-target indel mutation. (A) Cas9 protein levels. (B) On-target indel frequencies at VEGFA1 site. (C) Off-target indel frequencies at OT-1 site. (D) Off-target indel frequencies at OT-2 site. HEK293 cells were co-transfected as in Figure 2 and cells were harvested at 0, 6, 12, 24, 48 and 72 h. Cas9 levels were analyzed by Western blot and scanning densitometry. The Cas9 expression levels were normalized to that of the internal control and then normalized to that in the CLY/gV group at 24 h (defined as 100%) to yield the relative Cas9 protein expression levels (%). The indel frequencies at VEGFA1, OT-1 and OT-2 sites were analyzed as in Figure 3. The data represent the averages ±SD of two independent culture experiments.
Despite the stringently suppressed Cas9 expression, the K-CLY/gVgC group resulted in an on-target mutation kinetics similar to other groups, with a high on-target indel frequency (20.0 ± 0.8%) at 6 h, which slightly increased to 26.5 ± 1.1% at 24 h and barely increased thereafter (Figure 5B), confirming that a low level, transient Cas9 expression was sufficient to trigger effective on-target mutation at as early as 6 h.
Furthermore, uncontrolled Cas9 expression in the CLY/gV group led to increasing off-target mutation over time, with the indel frequency at OT-1 rising from 16.3 ± 0.2% at 24 h to 22.8 ± 0.1% at 72 h (Figure 5C) and the frequency at OT-2 elevating from 9.5 ± 0.2% at 24 h to 17.1 ± 1.1% at 72 h (Figure 5D). Restricting the Cas9 expression by self-cleavage (CLY/gVgC group) or by L7Ae:K-turn repression (K-CLY/gV group) decreased the off-target mutation frequencies to ≈10.5–11.5% at OT-1 and ≈6.1–6.3% at OT-2 at 24 h, and the off-target frequencies slightly increased after 24 h. Using the synthetic switch (K-CLY/gVgC group), the indel frequency dropped to 4.7±0.4% at 24 h and barely increased thereafter at OT-1 (Figure 5C). The indel frequency remained lower than 3% through 72 h at OT-2 (Figure 5D). These data highlighted the importance of transient and low level expression of Cas9 to minimize off-target effects.
DISCUSSION
It is well documented that uncontrolled and excessive Cas9 expression, mediated by either viral or nonviral vector, induces high frequency off-target effects (for review see (24,25)). To tackle this problem, different strategies to control Cas9 expression or activity have been developed. For instance, Cas9 expression can be driven by an inducible promoter (44,45) or Cas9 can be expressed in a split or inactive form (46–51). Consequently, Cas9 can be expressed or activated only upon stimulation with a chemical inducer (44,45,52,53) or by exposure to light (51,54). These approaches indeed effectively control Cas9 expression/activity temporally and reduce the off-target effects. However, inducible promoters often suffer from leaky expression while chemical inducers may elicit cytotoxicity and make in vivo genome editing more complicated. Conversely, the light-induction approach may be limited to in vitro applications because stimulating Cas9 expression/activity in vivo with light requires more invasive equipment and light penetration into deep tissue may be problematic (25).
Inspired by synthetic biology, here we built a genetic switch that refrains Cas9 expression not only in the transcription step by self-cleavage of cas9, but also in the translation step by L7Ae:K-turn system (Figure 1). Such synthetic switch enables self-regulation and obviates the need of external stimuli for activation. In agreement with common notions, uncontrolled Cas9 expression (CLY/gV group) gave rise to robust and excessive Cas9 expression, which induced high on-target mutagenesis and overt off-target effects in HEK293, HeLa and HepG2 cells at 24 h post-transfection (Figures 2–4). Using the gRNA-mediated self-cleavage approach alone (CLY/gVgC group), Cas9 expression was reduced for only ≈40% (Figure 2D). The off-target effects were diminished, but remained evident (Figures 3 and 4). Translational repression alone using the L7Ae:K-turn system (K-CLY/gV group) further reduced the Cas9 expression for ≈90% (Figure 2D). However, the off-target effects were not further abolished in all cell lines tested (Figures 3 and 4), indicating a Cas9 concentration range in which lowering Cas9 expression level does not further improve the specificity. Nonetheless, the synthetic switch exploiting both cas9 self-destruction and L7Ae:K-turn (K-CLY/gVgC group) significantly attenuated the Cas9 expression for ≈98% (Figure 2D). Such low level Cas9 expression potentiated the minimization of off-target effects, without compromising the on-target mutagenesis (Figures 3 and 4 and Supplementary Figures S2–S4). Furthermore, we employed two web-based tools (TagScan (12) and COSMID (55)) to predict potential off-target sites resulting from the gRNA targeting cas9, and found no meaningful off-targets, suggesting that the cas9-targeting gRNA induced no additional off-target effects.
Another intriguing finding is the correlation between Cas9 expression kinetics and the on-target/off-target mutagenesis (Figure 5). We found detectable Cas9 expression and concomitant high frequency on-target mutagenesis at as early as 6 h in all groups (Figure 5). Without simultaneous repression in the translation and transcription steps (CLY/gV, CLY/gVgC and K-CLY/gV groups), the Cas9 expression continued to rise with concurrent increase of off-target effects, but the on-target mutagenesis frequency did not markedly increase. In contrast, the synthetic switch regulation (K-CLY/gVgC group) limited the Cas9 expression and off-target effects to minimal levels through 72 h. These results demonstrated that a very low level Cas9 expression is sufficient to induce on-target mutagenesis in a short period of time. Moreover, persistent presence/expression of Cas9 does not markedly increase the on-target mutagenesis. These findings underscore the importance of pulse Cas9 expression to minimize the off-target effects, and echo the notion that Cas9 induces indels very shortly in 3 h and reaches a plateau by 24 h, as discovered in a study using RNP transfection (17). In our synthetic switch design, the residual Cas9 still induced slightly increased off-target effects at 72 h (Figure 5). This problem may be tackled by fusing a destabilizing peptide (DD) to Cas9 as described recently so as to shorten the half-life of Cas9 (56) and further restrict the occurrence of off-target mutagenesis.
In addition to restricting the Cas9 expression, off-target effects may be reduced by truncating the gRNA sequence (57), chemically modifying the gRNA (58), using Cas9 nickase (59) or modifying Cas9 nuclease activity through the protein engineering approach (37,38). Although the engineered Cas9 (e.g. eSpCas9 (37) or Cas9-HF1 (38)) effectively reduces the off-target effects, the cognate PAM sequence (NGG) for Cas9 may not be present in the close proximity of the desired sequence to edit. As such, a growing number of other Cas9 orthologues with different requirements and specificity in PAM sequences have been exploited for genome engineering (8). Most of these Cas9 orthologues have not been optimized to reduce off-target effects as the protein engineering approach requires intensive mutagenesis and time-consuming screening (8). Our synthetic switch strategy may be exploited to drive low level, self-regulated expression of these orthologous Cas9 nucleases, in combination with optimized gRNA design (e.g. truncating the gRNA seed sequence to 17 nt), for precise genome engineering with minimal off-target effects.
Most importantly, viral vectors such as LV, integrase-deficient lentivirus (IDLV), adenovirus (Ad) and adeno-associated virus (AAV) have been utilized to deliver the CRISPR/Cas9 system in vitro or in vivo (26,48,60,61). These viral vectors enable efficient gene delivery and are generally more effective than RNP or non-viral vectors for in vivo genome engineering (62). In particular, AAV has been approved for gene therapy and is the most widely used vector for CRISPR/Cas9 delivery (48,63,64). However, the packaging capacity for these viral vectors (>8 kb for Ad, ≈8 kb for IDLV and ≈4.7–5 kb for AAV) is limited (65), which often hinders the incorporation of regulatory elements into the viral vectors. Without appropriate regulation, the Cas9 expression could last for weeks to months (26), hence inducing high frequency off-target effects. Furthermore, Cas9 expression was found to induce immune responses in vivo (48,66) and a recent study uncovered pre-existing adaptive immunity to Cas9 protein in humans (https://www.biorxiv.org/content/early/2018/01/05/243345). These studies highlight the importance of attenuating and restricting Cas9 expression to minimize potential side effects. The genetic device we designed, including the complete CMV promoter/enhancer (584 bp), two repeats of K-turn motifs (53 bp), NLS (21 bp), Cas9 (4101 bp), L7Ae (357 bp), EYFP (720 bp), two P2A sequences (66 bp each) and poly A signal (225 bp), is only 6193 bp in size (Supplementary Figure S6A). This device is compact enough to be packaged into IDLV and Ad, and can be used together with another gRNA expression viral vector for genome engineering. By using a smaller truncated CMV promoter (173 bp), a minimal poly A signal (49 bp) and removing the EYFP reporter, the device can be downsized to 4820 bp (Supplementary Figure S6B). The size of the device may be further reduced to <4000 bp (Supplementary Figure S6C) by using a smaller Cas9 orthologue such as SaCas9 (≈3156 bp (40)) or CjCas9 (2952 bp (67)), thus allowing for packaging into the most widely used AAV vector for CRISPR-mediated in vivo genome editing (62). Such AAV vector can be applied in combination with another AAV expressing the gRNA and can simultaneously self-regulate the expression of Cas9 and L7Ae by transcriptional and translational inhibition, hence reducing the possible side effects caused by Cas9 and L7Ae expression.
In summary, here we engineered a synthetic switch for stringent control of Cas9 expression in both transcription and translation steps to minimize the off-target effects without sacrificing the on-target mutagenesis frequency. We also uncovered that the synthetic switch-regulated Cas9 expression can induce high on-target mutation frequency at as early as 6 h while restricting the off-target effects to a minimal level through 72 h. Such synthetic switch design provides a novel ‘hit and run’ strategy and may be incorporated into viral vectors to enhance the safety profile for in vivo genome engineering.
Supplementary Material
ACKNOWLEDGEMENTS
Author contributions: C.C.S. conducted the experiments and wrote the paper. M.N.H. conducted the experiments. C.W.C. conducted experiments and wrote the paper. M.W.L. conducted the experiments. J.R.H. designed the project. Y.T. conducted the experiments. Y.C.H. designed the project and wrote the paper.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Ministry of Science and Technology (MOST) [106-2622-E-007-014-CC1, 105-2923-E-007-002-MY3, 106-2221-E-007-085-MY3, 107-3017-F-007-002, 107-2622-E-007-003-CC1]; Frontier Research Center on Fundamental and Applied Sciences of Matters; Featured Areas Research Center Program within the framework of the Higher Education Sprout Project by the Ministry of Education (MOE) [107QR001I5, 107Q2529E1], Taiwan. Funding for open access charge: MOST [107-2622-E-007-003-CC1].
Conflict of interest statement. None declared.
REFERENCES
- 1. Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E.. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012; 337:816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A. et al. . Multiplex genome engineering using CRISPR/Cas systems. Science. 2013; 339:819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mali P., Yang L., Esvelt K.M., Aach J., Guell M., DiCarlo J.E., Norville J.E., Church G.M.. RNA-guided human genome engineering via Cas9. Science. 2013; 339:823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hsu P.D., Lander E.S., Zhang F.. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014; 157:1262–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chung M.E., Yeh I.H., Sung L.Y., Wu M.Y., Chao Y.P., Ng I.S., Hu Y.C.. Enhanced integration of large DNA into E. coli chromosome by CRISPR/Cas9. Biotechnol. Bioeng. 2017; 114:172–183. [DOI] [PubMed] [Google Scholar]
- 6. Li H., Shen C.R., Huang C.-H., Sung L.-Y., Wu M.-Y., Hu Y.-C.. CRISPR-Cas9 for the genome engineering of cyanobacteria and succinate production. Metab. Eng. 2016; 38:293–302. [DOI] [PubMed] [Google Scholar]
- 7. Wright A.V., Nunez J.K., Doudna J.A.. Biology and applications of CRISPR systems: harnessing nature's toolbox for genome engineering. Cell. 2016; 164:29–44. [DOI] [PubMed] [Google Scholar]
- 8. Komor A.C., Badran A.H., Liu D.R.. CRISPR-based technologies for the manipulation of eukaryotic genomes. Cell. 2017; 168:20–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Maeder M.L., Gersbach C.A.. Genome-editing technologies for gene and cell therapy. Mol. Ther. 2016; 24:430–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Muller M., Lee C.M., Gasiunas G., Davis T.H., Cradick T.J., Siksnys V., Bao G., Cathomen T., Mussolino C.. Streptococcus thermophilus CRISPR-Cas9 systems enable specific editing of the human genome. Mol. Ther. 2016; 24:636–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fu Y., Foden J.A., Khayter C., Maeder M.L., Reyon D., Joung J.K., Sander J.D.. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat. Biotechnol. 2013; 31:822–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lin Y., Cradick T.J., Brown M.T., Deshmukh H., Ranjan P., Sarode N., Wile B.M., Vertino P.M., Stewart F.J., Bao G.. CRISPR/Cas9 systems have off-target activity with insertions or deletions between target DNA and guide RNA sequences. Nucleic Acids Res. 2014; 42:7473–7485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cho S.W., Kim S., Kim J.M., Kim J.S.. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat. Biotechnol. 2013; 31:230–232. [DOI] [PubMed] [Google Scholar]
- 14. Zhang Y., Liang Z., Zong Y., Wang Y., Liu J., Chen K., Qiu J.-L., Gao C.. Efficient and transgene-free genome editing in wheat through transient expression of CRISPR/Cas9 DNA or RNA. Nat. Commun. 2016; 7:12617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yin H., Song C.-Q., Dorkin J.R., Zhu L.J., Li Y., Wu Q., Park A., Yang J., Suresh S., Bizhanova A. et al. . Therapeutic genome editing by combined viral and non-viral delivery of CRISPR system components in vivo. Nat. Biotechnol. 2016; 34:328–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Carlson-Stevermer J., Abdeen A.A., Kohlenberg L., Goedland M., Molugu K., Lou M., Saha K.. Assembly of CRISPR ribonucleoproteins with biotinylated oligonucleotides via an RNA aptamer for precise gene editing. Nat. Commun. 2017; 8:1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kim S., Kim D., Cho S.W., Kim J., Kim J.-S.. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res. 2014; 24:1012–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Svitashev S., Schwartz C., Lenderts B., Young J.K., Mark Cigan A.. Genome editing in maize directed by CRISPR–Cas9 ribonucleoprotein complexes. Nat. Commun. 2016; 7:13274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Staahl B.T., Benekareddy M., Coulon-Bainier C., Banfal A.A., Floor S.N., Sabo J.K., Urnes C., Munares G.A., Ghosh A., Doudna J.A.. Efficient genome editing in the mouse brain by local delivery of engineered Cas9 ribonucleoprotein complexes. Nat. Biotechnol. 2017; 35:431–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zuris J.A., Thompson D.B., Shu Y., Guilinger J.P., Bessen J.L., Hu J.H., Maeder M.L., Joung J.K., Chen Z.-Y., Liu D.R.. Cationic lipid-mediated delivery of proteins enables efficient protein-based genome editing in vitro and in vivo. Nat. Biotechnol. 2014; 33:73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lee K., Conboy M., Park H.M., Jiang F., Kim H.J., Dewitt M.A., Mackley V.A., Chang K., Rao A., Skinner C. et al. . Nanoparticle delivery of Cas9 ribonucleoprotein and donor DNA in vivo induces homology-directed DNA repair. Nat. Biomed. Eng. 2017; 1:889–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kaczmarek J.C., Kowalski P.S., Anderson D.G.. Advances in the delivery of RNA therapeutics: from concept to clinical reality. Genome Med. 2017; 9:60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mout R., Ray M., Lee Y.-W., Scaletti F., Rotello V.M.. In vivo delivery of CRISPR/Cas9 for therapeutic gene editing: Progress and challenges. Bioconj. Chem. 2017; 28:880–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tsai S.Q., Joung J.K.. Defining and improving the genome-wide specificities of CRISPR-Cas9 nucleases. Nat. Rev. Genet. 2016; 17:300–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chira S., Gulei D., Hajitou A., Zimta A.A., Cordelier P., Berindan-Neagoe I.. CRISPR/Cas9: Transcending the reality of genome editing. Mol. Ther. Nucleic Acids. 2017; 7:211–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chen Y., Liu X., Zhang Y., Wang H., Ying H., Liu M., Li D., Lui K.O., Ding Q.. A self-restricted CRISPR system to reduce off-target effects. Mol. Ther. 2016; 24:1508–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Petris G., Casini A., Montagna C., Lorenzin F., Prandi D., Romanel A., Zasso J., Conti L., Demichelis F., Cereseto A.. Hit and go Cas9 delivered through a lentiviral based self-limiting circuit. Nat. Commun. 2017; 8:15334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ruan G.X., Barry E., Yu D., Lukason M., Cheng S.H., Scaria A.. CRISPR/Cas9-mediated genome editing as a therapeutic approach for Leber Congenital Amaurosis 10. Mol. Ther. 2017; 25:331–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Smole A., Lainšček D., Bezeljak U., Horvat S., Jerala R.. A synthetic mammalian therapeutic gene circuit for sensing and suppressing inflammation. Mol. Ther. 2017; 25:102–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ausländer S., Fussenegger M.. Synthetic RNA-based switches for mammalian gene expression control. Curr. Opin. Biotechnol. 2017; 48:54–60. [DOI] [PubMed] [Google Scholar]
- 31. Saito H., Kobayashi T., Hara T., Fujita Y., Hayashi K., Furushima R., Inoue T.. Synthetic translational regulation by an L7Ae–kink-turn RNP switch. Nat. Chem. Biol. 2010; 6:71–78. [DOI] [PubMed] [Google Scholar]
- 32. Saito H., Fujita Y., Kashida S., Hayashi K., Inoue T.. Synthetic human cell fate regulation by protein-driven RNA switches. Nat. Commun. 2011; 2:160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Endo K., Stapleton J.A., Hayashi K., Saito H., Inoue T.. Quantitative and simultaneous translational control of distinct mammalian mRNAs. Nucleic Acids Res. 2013; 41:e135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Stapleton J.A., Endo K., Fujita Y., Hayashi K., Takinoue M., Saito H., Inoue T.. Feedback control of protein expression in mammalian cells by tunable synthetic translational inhibition. ACS Syn. Biol. 2012; 1:83–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Auslander S., Stucheli P., Rehm C., Auslander D., Hartig J.S., Fussenegger M.. A general design strategy for protein-responsive riboswitches in mammalian cells. Nat. Meth. 2014; 11:1154–1160. [DOI] [PubMed] [Google Scholar]
- 36. Wroblewska L., Kitada T., Endo K., Siciliano V., Stillo B., Saito H., Weiss R.. Mammalian synthetic circuits with RNA binding proteins for RNA-only delivery. Nat. Biotechnol. 2015; 33:839–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Slaymaker I.M., Gao L., Zetsche B., Scott D.A., Yan W.X., Zhang F.. Rationally engineered Cas9 nucleases with improved specificity. Science. 2016; 351:84–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kleinstiver B.P., Pattanayak V., Prew M.S., Tsai S.Q., Nguyen N.T., Zheng Z., Joung J.K.. High-fidelity CRISPR–Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 2016; 529:490–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kleinstiver B.P., Prew M.S., Tsai S.Q., Topkar V.V., Nguyen N.T., Zheng Z., Gonzales A.P.W., Li Z., Peterson R.T., Yeh J.-R.J. et al. . Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature. 2015; 523:481–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ran F.A., Cong L., Yan W.X., Scott D.A., Gootenberg J.S., Kriz A.J., Zetsche B., Shalem O., Wu X., Makarova K.S. et al. . In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015; 520:186–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gabriel R., von Kalle C., Schmidt M.. Mapping the precision of genome editing. Nat. Biotechnol. 2015; 33:150. [DOI] [PubMed] [Google Scholar]
- 42. Zhang X.H., Tee L.Y., Wang X.G., Huang Q.S., Yang S.H.. Off-target effects in CRISPR/Cas9-mediated genome engineering. Mol. Ther. Nucleic Acids. 2015; 4:e264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Brinkman E.K., Chen T., Amendola M., van Steensel B.. Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res. 2014; 42:e168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Cao J., Wu L., Zhang S.M., Lu M., Cheung W.K., Cai W., Gale M., Xu Q., Yan Q.. An easy and efficient inducible CRISPR/Cas9 platform with improved specificity for multiple gene targeting. Nucleic Acids Res. 2016; 44:e149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Dow L.E., Fisher J., O’Rourke K.P., Muley A., Kastenhuber E.R., Livshits G., Tschaharganeh D.F., Socci N.D., Lowe S.W.. Inducible in vivo genome editing with CRISPR-Cas9. Nat. Biotechnol. 2015; 33:390–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Truong D.-J.J., Kühner K., Kühn R., Werfel S., Engelhardt S., Wurst W., Ortiz O.. Development of an intein-mediated split–Cas9 system for gene therapy. Nucleic Acids Res. 2015; 43:6450–6458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Davis K.M., Pattanayak V., Thompson D.B., Zuris J.A., Liu D.R.. Small molecule–triggered Cas9 protein with improved genome-editing specificity. Nat. Chem. Biol. 2015; 11:316–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Chew W.L., Tabebordbar M., Cheng J.K.W., Mali P., Wu E.Y., Ng A.H.M., Zhu K., Wagers A.J., Church G.M.. A multifunctional AAV–CRISPR–Cas9 and its host response. Nat. Meth. 2016; 13:868–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tsai S.Q., Wyvekens N., Khayter C., Foden J.A., Thapar V., Reyon D., Goodwin M.J., Aryee M.J., Joung J.K.. Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nat. Biotechnol. 2014; 32:569–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Guilinger J.P., Thompson D.B., Liu D.R.. Fusion of catalytically inactive Cas9 to FokI nuclease improves the specificity of genome modification. Nat. Biotechnol. 2014; 32:577–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Nihongaki Y., Kawano F., Nakajima T., Sato M.. Photoactivatable CRISPR-Cas9 for optogenetic genome editing. Nat. Biotechnol. 2015; 33:755–760. [DOI] [PubMed] [Google Scholar]
- 52. Liu K.I., Ramli M.N.B., Woo C.W.A., Wang Y., Zhao T., Zhang X., Yim G.R.D., Chong B.Y., Gowher A., Chua M.Z.H. et al. . A chemical-inducible CRISPR–Cas9 system for rapid control of genome editing. Nat. Chem. Biol. 2016; 12:980–987. [DOI] [PubMed] [Google Scholar]
- 53. Lu J., Zhao C., Zhao Y., Zhang J., Zhang Y., Chen L., Han Q., Ying Y., Peng S., Ai R. et al. . Multimode drug inducible CRISPR/Cas9 devices for transcriptional activation and genome editing. Nucleic Acids Res. 2017; 46:e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zhou X.X., Zou X., Chung H.K., Gao Y., Liu Y., Qi L.S., Lin M.Z.. A single-chain photoswitchable CRISPR-Cas9 architecture for light-inducible gene editing and transcription. ACS Chem. Biol. 2018; 13:443–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Cradick T.J., Qiu P., Lee C.M., Fine E.J., Bao G.. COSMID: A web-based tool for identifying and validating CRISPR/Cas off-target sites. Mol. Ther.-Nucleic Acids. 2014; 3:e214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Senturk S., Shirole N.H., Nowak D.G., Corbo V., Pal D., Vaughan A., Tuveson D.A., Trotman L.C., Kinney J.B., Sordella R.. Rapid and tunable method to temporally control gene editing based on conditional Cas9 stabilization. Nat. Commun. 2017; 8:14370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Fu Y., Sander J.D., Reyon D., Cascio V.M., Joung J.K.. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat. Biotechnol. 2014; 32:279–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ryan D.E., Taussig D., Steinfeld I., Phadnis S.M., Lunstad B.D., Singh M., Vuong X., Okochi K.D., McCaffrey R., Olesiak M. et al. . Improving CRISPR–Cas specificity with chemical modifications in single-guide RNAs. Nucleic Acids Res. 2018; 46:792–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Ran F.A., Hsu Patrick D., Lin C.-Y., Gootenberg Jonathan S., Konermann S., Trevino A.E., Scott David A., Inoue A., Matoba S., Zhang Y. et al. . Double nicking by RNA-Guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013; 154:1380–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ortinski P.I., O’Donovan B., Dong X.Y., Kantor B.. Integrase-deficient lentiviral vector as an all-in-one platform for highly efficient CRISPR/Cas9-mediated gene editing. Mol. Ther.-Meth. Clin. Dev. 2017; 5:153–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Holkers M., Maggio I., Henriques S.F.D., Janssen J.M., Cathomen T., Goncalves M.A.F.V.. Adenoviral vector DNA for accurate genome editing with engineered nucleases. Nat. Meth. 2014; 11:1051–1057. [DOI] [PubMed] [Google Scholar]
- 62. Chen X., Goncalves M.A.F.V.. Engineered viruses as genome editing devices. Mol. Ther. 2016; 24:447–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Lau C.-H., Suh Y.. In vivo genome editing in animals using AAV-CRISPR system: applications to translational research of human disease [version 1; referees: 2 approved]. F1000Res. 2017; 6:2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Yin H., Song C.-Q., Dorkin J.R., Zhu L.J., Li Y., Wu Q., Park A., Yang J., Suresh S., Bizhanova A. et al. . Therapeutic genome editing by combined viral and non-viral delivery of CRISPR system components in vivo. Nat. Biotechnol. 2016; 34:328–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Wu Z.J., Yang H.Y., Colosi P.. Effect of genome size on AAV vector packaging. Mol. Ther. 2010; 18:80–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Leong C.W. Immunity to CRISPR Cas9 and Cas12a therapeutics. Wiley Interdiscip. Rev. Syst. Biol. Med. 2018; 10:e1408. [DOI] [PubMed] [Google Scholar]
- 67. Kim E., Koo T., Park S.W., Kim D., Kim K., Cho H.Y., Song D.W., Lee K.J., Jung M.H., Kim S. et al. . In vivo genome editing with a small Cas9 orthologue derived from Campylobacter jejuni. Nat. Commun. 2017; 8:14500. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.