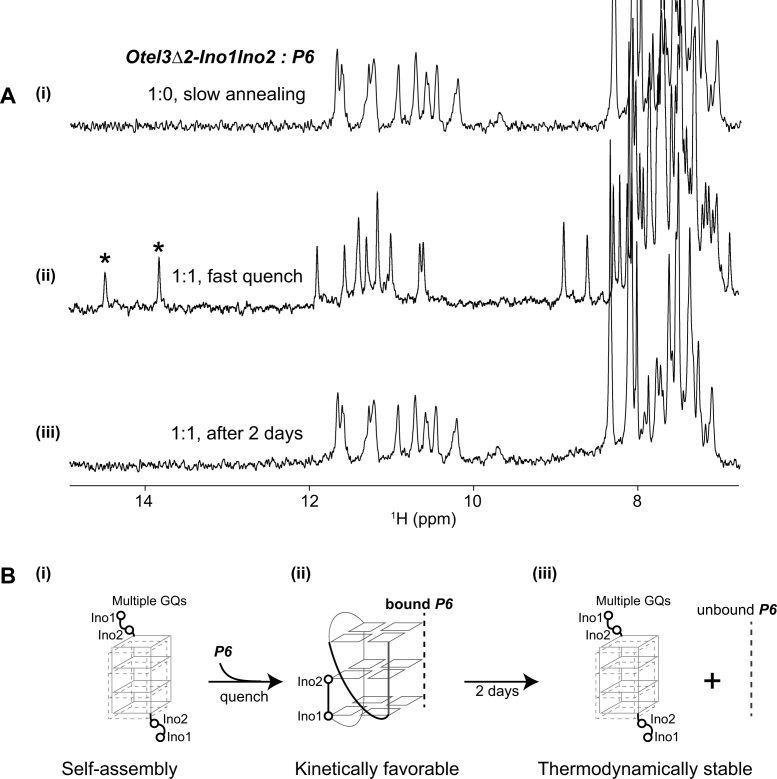
Figure 6.
The dual inosine-substitution mutant Otel3Δ2-Ino1Ino2 probed by the short chain P6. (A) (i) The one-dimensional 1H spectrum of Otel3Δ2-Ino1Ino2 alone showed its self-assembly into multiple G-quadruplexes under annealing condition. (ii) The one-dimensional 1H spectrum of the Otel3Δ2-Ino1Ino2/P6 complex at an equimolar ratio under quench conditions showed the formation of a new G-quadruplex containing two hydrogen-bonded inosine imino peaks (marked by asterisks). (iii) The one-dimensional 1H spectrum of the sample used in panel (ii) after incubation at room temperature for 2 days. The concentration of the DNA strands was 0.1 mM. The possible process of conformational changes under the conditions shown in panel (A) was schematically indicated using the structural model in panel (B), without showing the unfolding proportions of single chain P6 and single chain Otel3Δ2-Ino1Ino2 for clarity. (B) (i) and (iii) These two inosines did not participate in the formation of the G-tetrad core. In the schematic structure, the short chain P6 is indicated by the gray dotted line. The hollow circles represent the 5′ terminal inosines of Otel3Δ2-Ino1Ino2.