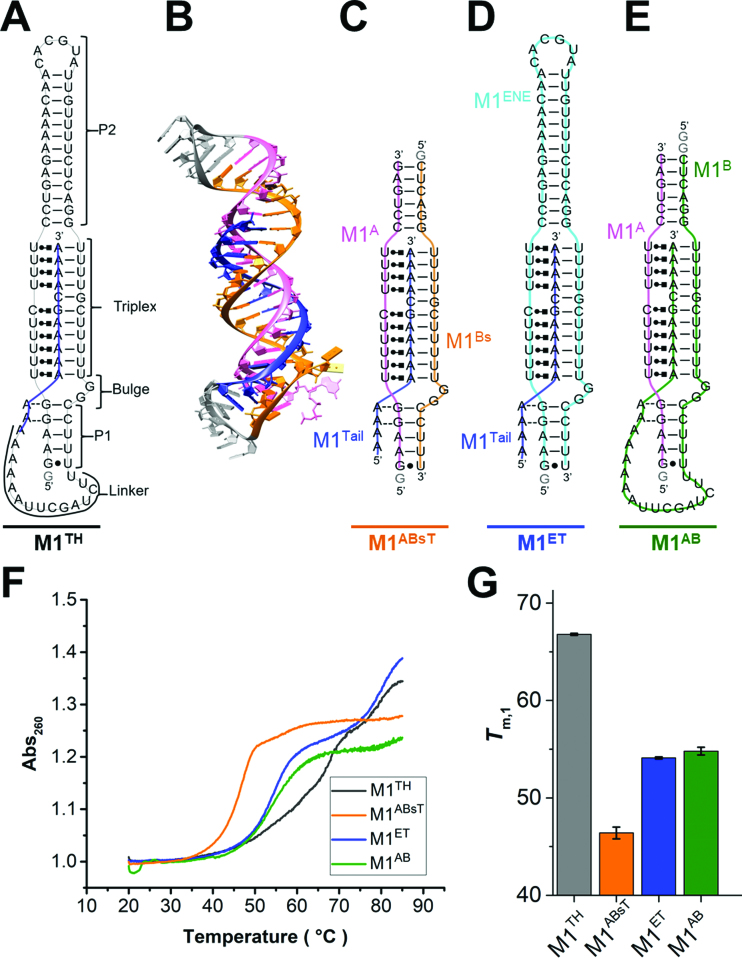
Figure 1.
Peripheral elements regulate thermostability of the MALAT1 ENE triple helix. (A) Secondary structure of the full-length MALAT1 ENE triple helix (M1TH) including Watson–Crick interactions (—), Hoogsteen interactions ( ), A-minor interactions (−−), and a wobble base pair (•). A 5′ GTP (grey) is added to the sequence to facilitate transcription. The A-rich tail is colored blue and structural regions are indicated in brackets. (B) The crystal structure of the MALAT1 triple helix core (PDB: 4PLX) (33). The three strands are colored orange, pink, and blue. The two short loops (gray) were engineered to facilitate crystallization. (C) Secondary structure of trimolecular M1ABsT. M1Bs is colored orange, M1A is colored pink, and M1Tail is colored blue, as in B. (D) Secondary structure of bimolecular M1ET, which eliminates the basal linker. The ENE motif is colored cyan and the M1Tail is colored as in C. (E) Secondary structure of bimolecular M1AB, wherein the basal linker is included and the apical P2 helix is truncated. M1A is colored as in C and M1B is colored green. In C–E, a 5′ GTP (gray) is added to the sequence to facilitate transcription. (F) Normalized UV melt profiles for M1TH (gray), M1ABsT (orange), M1ET (blue), M1AB (green) from 20 to 85°C in 20 mM HEPES•KOH, pH 7.4, 1 mM MgCl2, 25 mM NaCl and 25 mM KCl. Triplex formation for all constructs is explicitly demonstrated in Supplementary Figure S2. Triplex and duplex melting occur in the same transition for the bimolecular M1AB. All experiments were performed in duplicate. A single representative melting profile is plotted. (G) Triplex melting temperatures (Tm,1) for the unimolecular, bimolecular, and trimolecular constructs, colored as in F. Large changes in Tm,1 for the bi- and tri-molecular RNA relative to the full-length RNA demonstrate the contributions of peripheral elements to triplex stability.
), A-minor interactions (−−), and a wobble base pair (•). A 5′ GTP (grey) is added to the sequence to facilitate transcription. The A-rich tail is colored blue and structural regions are indicated in brackets. (B) The crystal structure of the MALAT1 triple helix core (PDB: 4PLX) (33). The three strands are colored orange, pink, and blue. The two short loops (gray) were engineered to facilitate crystallization. (C) Secondary structure of trimolecular M1ABsT. M1Bs is colored orange, M1A is colored pink, and M1Tail is colored blue, as in B. (D) Secondary structure of bimolecular M1ET, which eliminates the basal linker. The ENE motif is colored cyan and the M1Tail is colored as in C. (E) Secondary structure of bimolecular M1AB, wherein the basal linker is included and the apical P2 helix is truncated. M1A is colored as in C and M1B is colored green. In C–E, a 5′ GTP (gray) is added to the sequence to facilitate transcription. (F) Normalized UV melt profiles for M1TH (gray), M1ABsT (orange), M1ET (blue), M1AB (green) from 20 to 85°C in 20 mM HEPES•KOH, pH 7.4, 1 mM MgCl2, 25 mM NaCl and 25 mM KCl. Triplex formation for all constructs is explicitly demonstrated in Supplementary Figure S2. Triplex and duplex melting occur in the same transition for the bimolecular M1AB. All experiments were performed in duplicate. A single representative melting profile is plotted. (G) Triplex melting temperatures (Tm,1) for the unimolecular, bimolecular, and trimolecular constructs, colored as in F. Large changes in Tm,1 for the bi- and tri-molecular RNA relative to the full-length RNA demonstrate the contributions of peripheral elements to triplex stability.