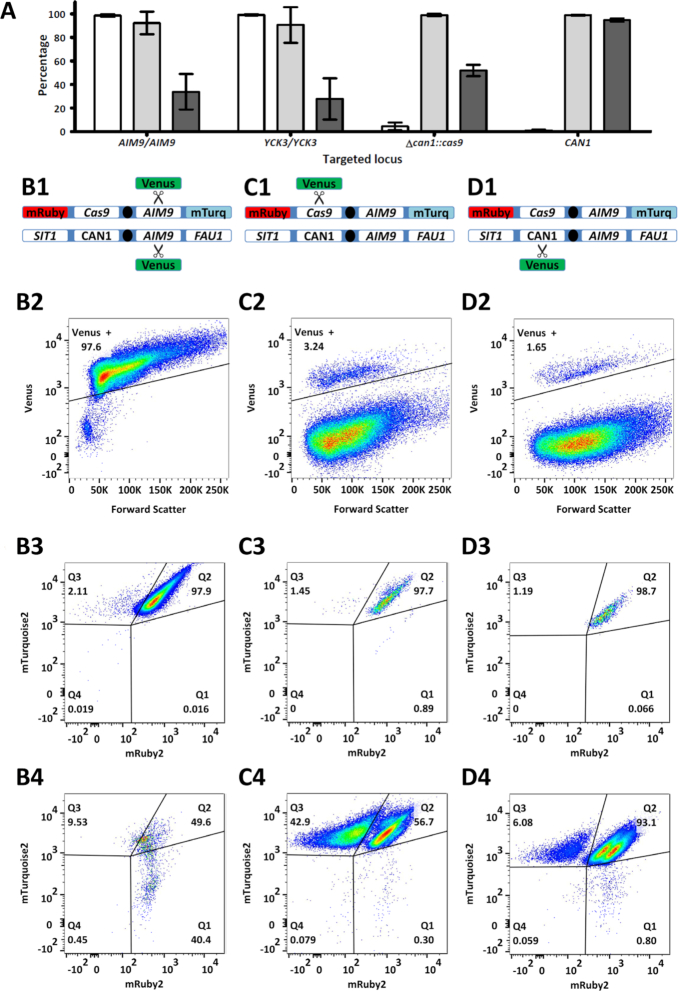
Figure 2.
Cas9-mediated gene editing of homozygous and heterozygous loci on chromosome V of S. cerevisiae. (A) Average fluorescence of cell populations in which the homozygous AIM9 and YCK3 alleles and the heterozygous cas9 and CAN1 alleles were targeted in the diploid strain IMX1557. The percentage of cells expressing Venus (white), the percentages of cells expressing both mTurquoise2 and mRuby2 in the Venus positive (light gray) and in Venus negative cells (dark gray) are shown. For each target, averages and standard deviation for biological triplicates are shown. When targeting the hemizygous CAN1 allele, LOH manifests itself in a lower percentage of cells expressing Venus, but it does not affect mTurquoise2 and mRuby2 fluorescence, as these fluorophores are located on the non-targeted chromosome copy. (B–D) Fluorescence profiles obtained when targeting AIM9, cas9 and CAN1 in IMX1557. (row 1) Schematic representation of both copies of chromosome V in IMX1557, with the alleles at the SIT1, CAN1, AIM9 and FAU1 loci and scissors indicating Cas9 targeting. While one chromosome copy has the wildtype alleles for all loci, the other copy has mRuby2 integrated in SIT1, cas9 integrated in CAN1 and mTurquoise2 integrated in FAU1. (rows 2, 3 and 4) Flow cytometry profiles of targeted cells. Each gene was targeted in three biological replicates and flow cytometric data for a representative replicate is shown. After transformation, 100 000 cells were analysed by flow cytometry and single cells were selected based on a FSC-A/FSC-H plot to avoid multicellular aggregates. For each replicate, at least 75 000 single cells remained and the fluorescence corresponding to Venus was used to determine gene-editing efficiency (row 2). For each gene, the fluorescence corresponding to mRuby2 and mTurquoise2 is plotted for the cells with expression of Venus (row 3) and for cells without expression of Venus (row 4). Fluorescence results for all samples are provided in Supplementary Table S1.