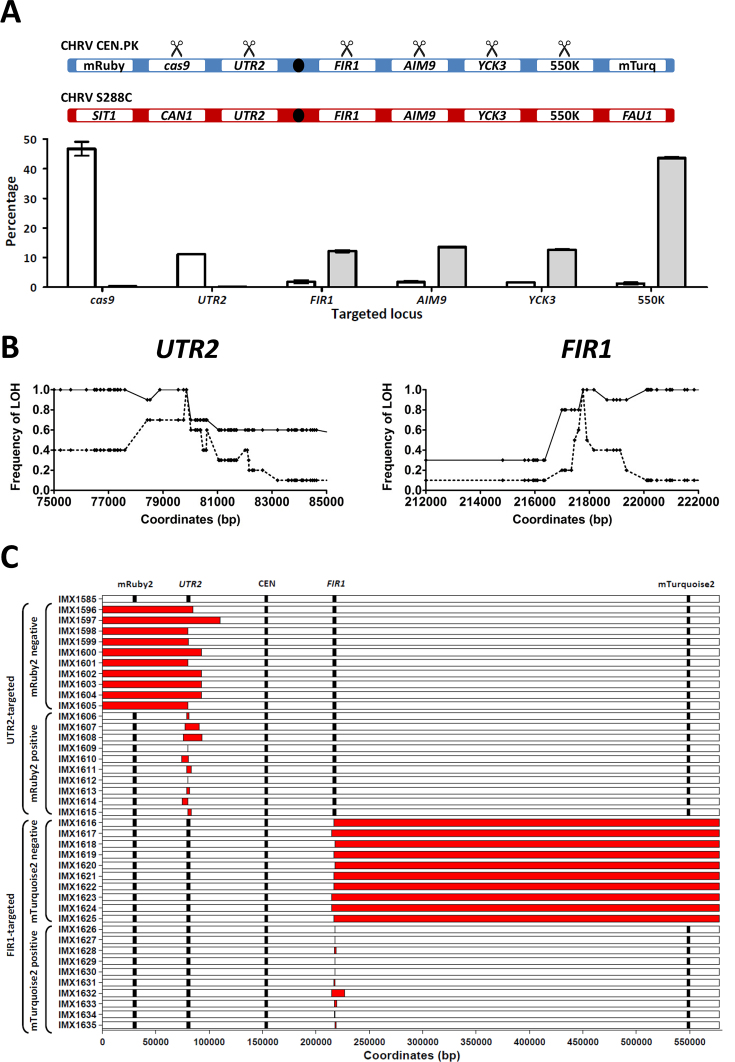
Figure 3.
Loss of heterozygosity caused by Cas9-mediated gene editing at heterozygous loci in the heterozygous S. cerevisiae diploid IMX1585. Mating of the haploid S. cerevisiae strains IMX1555 (CEN.PK-derived) and S288C yielded the heterozygous diploid strain IMX1585 (on average four heterozygous nucleotides per kbp on chromosome V). The CEN.PK-derived chromosome harbors the fluorophores mRuby2 and mTurquoise2, enabling detection of the loss of each arm of the CEN.PK-derived chromosome V by flow cytometry. DSBs were introduced specifically in the CEN.PK–derived chromosome and loss of heterozygosity was monitored at the population level using flow cytometry and in single cell isolates by whole genome sequencing. (A) Population-level loss of heterozygosity after targeting cas9, UTR2, FIR1, AIM9, YCK3 and 550K in IMX1585. The targeted loci on the CEN.PK-derived and S288C-derived chromosome V of IMX1585 are represented schematically. The SIT1, CAN1, UTR2, FIR2, AIM9, YCK3, 550K and FAU1 loci and the centromeres are indicated. The CEN.PK-derived chromosome harbors mRuby2 at the SIT1 locus and mTurquoise2 at the FAU1 locus. Scissors indicate CEN.PK-derived alleles which were specifically targeted using Cas9. In the graph, the percentage of cells having lost mRuby2 fluorescence (white) and mTurquoise2 (gray) is shown for each targeted locus. Averages and standard deviations were calculated from biological triplicates. (B) Loss of heterozygosity at the nucleotide level in single isolates obtained by targeting UTR2 and FIR1 in IMX1585. For both targeted loci, the frequency at which LOH was observed for each heterozygous nucleotide in a 10 kbp region around the targeted site was determined by whole genome sequencing. For UTR2, the 79,857th nucleotide was targeted and frequencies are indicated for 10 isolates with intact fluorescence (dashed line, isolates IMX1606-IMX1615) and 10 isolates having lost fluorescence corresponding to mRuby2 (continuous line, IMX1596-IMX1605). For FIR1, the 217,767th nucleotide was targeted and frequencies are indicated for 10 isolates with intact fluorescence (dashed line, isolates IMX1626-IMX1635) and 10 isolates having lost fluorescence corresponding to mTurquoise2 (continuous line, IMX1616-IMX1625). (C) Loss of heterozygosity at the chromosome scale in single isolates obtained by targeting UTR2 and FIR1 in IMX1585. Whole genome sequencing data was used to identify regions of chromosome V affected by loss of heterozygosity in isolates after targeting of UTR2 (IMX1596-IMX1615) and of FIR1 (IMX1616-IMX1635). For each strain, the fluorophores mRuby2 and mTurquoise2, the targeted genes UTR2 and FIR1 and the centromere are shown at their exact coordinates, but their size is not at scale. Loss of heterozygosity was defined as regions in which nucleotides which were heterozygous in IMX1585 were no longer heterozygous in the isolate (in red). In isolates which lost the fluorophore mRuby2 after targeting of UTR2 or which lost the fluorophore mTurquoise2 after targeting of FIR1, entire chromosome arms were affected by LOH. Exact coordinates are provided in Supplementary Table S2.