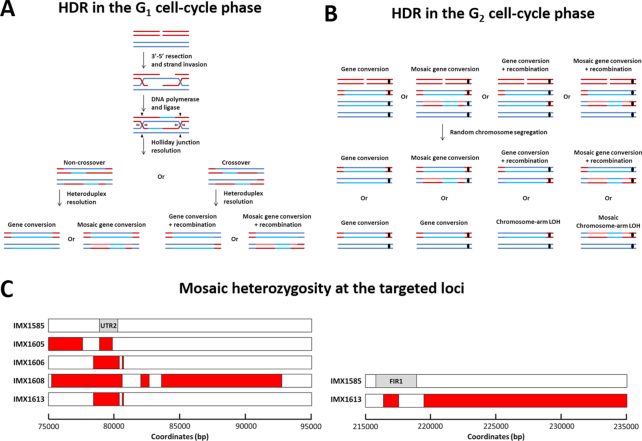
Figure 4.
Proposed mechanism for Cas9-mediated loss of heterozygosity in a diploid genome based on homologous recombination (HR) between homologous chromosomes. (A) Possible outcomes of HR in cells with one chromosome complement during the G1 stage of the cell cycle. (B) Possible outcomes of HR in cells with two chromosome complements during the G2 stage of the cell cycle. The targeted chromosome (red), its homolog (blue) and the centromere are indicated (black, where relevant). Newly synthesized DNA is shown in a lighter shade. Heteroduplex resolution occurs prior to chromatid segregation and the strand with the targeted NGG PAM sequence is always discarded due to Cas9 activity. For HR during the G2 stage of the cell cycle, HR occurs between one chromatid of the targeted and one chromatid of the non-targeted chromosome, as with HR during the G1 stage. The chromatids subsequently segregate according to their centromere pairing, with one red and one blue centromere in each daughter cell. For simplicity, only repair of one targeted chromatid is shown in the figure. Repair of both targeted sister chromatids results in the same genome alterations as shown here. As indicated in the figure, HR during the G2 stage of the cell cycle could yield local as well as chromosome-arm LOH by mitotic crossover, both with and without mosaic structures. (C) Mosaic loss of heterozygosity at the targeted loci in single isolates obtained by targeting UTR2 and FIR1 in IMX1585. Strains with mosaic loss of heterozygosity are at the UTR2 locus (left) and at the FIR1 locus (right) are indicated. The location of the ORFs is indicated in IMX1585 (gray). For each strain, heterozygous sequence is indicated in red. Exact coordinates of heterozygous nucleotides are indicated in Supplementary Table S2.