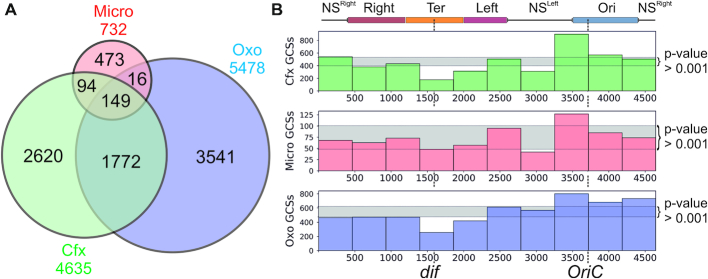
Figure 3.
Comparison of GCSs sets observed with Cfx, Micro, and Oxo. (A) Venn diagram representation of the relations between GCSs sets obtained with different gyrase poisons. (B) General distribution of poison-mediated GCSs revealed by Topo-Seq. E. coli W3110 MuSGS genome is divided into 10 non non-overlapping bins, the height of the bars indicates the number of GCSs in a particular genome region. Area between 0.0005 and 0.9995 quantile values (distribution of the number of GCSs in a bin set as binomial) are shown in light gray, so bars that fall out of this zone have a statistically significant deviation from a value expected for the uniform distribution. NSRight—right non-structured region; Right—right macrodomain; Ter—terminator domain; Left—left macrodomain; NSLeft – left non-structured region; Ori—origin domain. Macrodomains set as in (68).