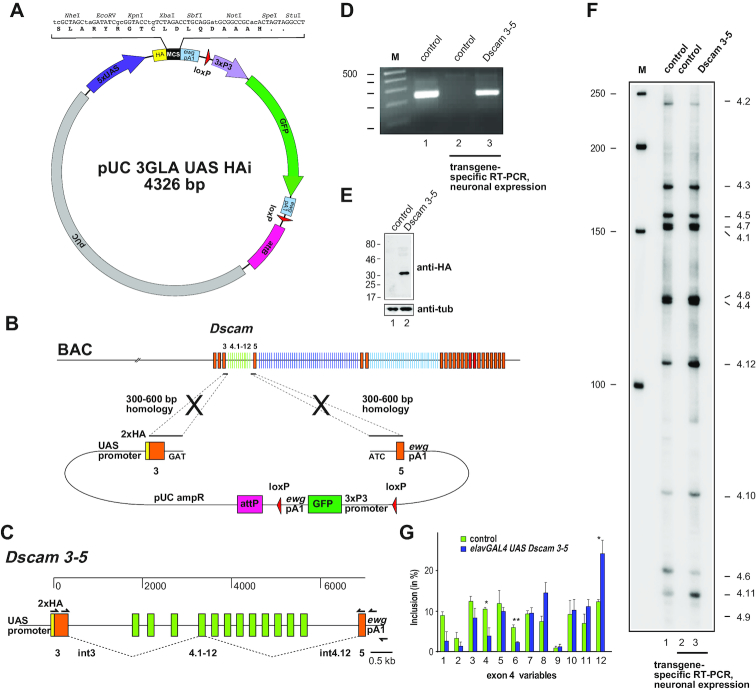
Figure 2.
A reporter transgene for the analysis of Dscam mutually exclusive alternative splicing. (A) Schematic of the pUC18 based high-copy number plasmid pUC 3GLA UAS HAi for Drosophila phiC31 transgenesis via the attB site (pink). The multiple cloning site (MCS, black) is shown on top indicating the reading frame for N-terminal tagging with a 3xhemagglutinin tag (HA, yellow). Heterologous expression is driven from the 5xUAS promoter (purple) by the GAL4 transcriptional activator and transcripts terminate at the polyA site from the ewg gene (light blue). The GFP marker (green) to identify transformed flies is driven by the artificial 3xP3 promoter (light purple) and flanked by loxP sites (red). (B) Schematic for the retrieval of the Dscam exon 3–5 region from a BAC containing the genomic locus using λ Red protein mediated gap-repair recombineering with a linearized plasmid containing homologous sequences of the beginning and the end of the construct. Homologous regions for recombination are indicated by crosses. (C) Graphical depiction of the Dscam exon 4 construct indicating primers for amplification of construct-specific transcripts using an RT primer in the ewg 3′UTR and nested PCR. (D) RT-PCR products for Dscam exon 4 shown on an agarose gel from the endogenous gene (lane 1) and from construct specific nested RT-PCR from control (lane 2) and transgenic flies with the Dscam 3–5 construct neuronally expressed with elavGAL4 (lane 3). (E) Western blot showing expression of the artificial Dscam 3–5 protein expressed with elavGAL4 in larval brains and detected with anti-HA antibodies (lane2), compared to wild type controls (lane 1). (F) Denaturing acrylamide gel showing inclusion of individual exon 4 variable exons from the endogenous gene (lane 1), and from construct specific nested RT-PCR from control (lane 2) and transgenic third instar larval brains from neuronal expression of the Dscam 3–5 construct with elavGAL4 (lane 3). Note that the endogenous Dscam gene is more broadly expressed and therefore shows slightly different inclusion levels for some exons. (G) Inclusion levels of exon 4 variables in larval brains of controls (green) and from transgenic expression of Dscam 3–5 from UAS in neurons using elavGAL4 (blue) shown as means with standard error from three experiments. Statistically significant differences are indicated above bars (*P < 0.05).