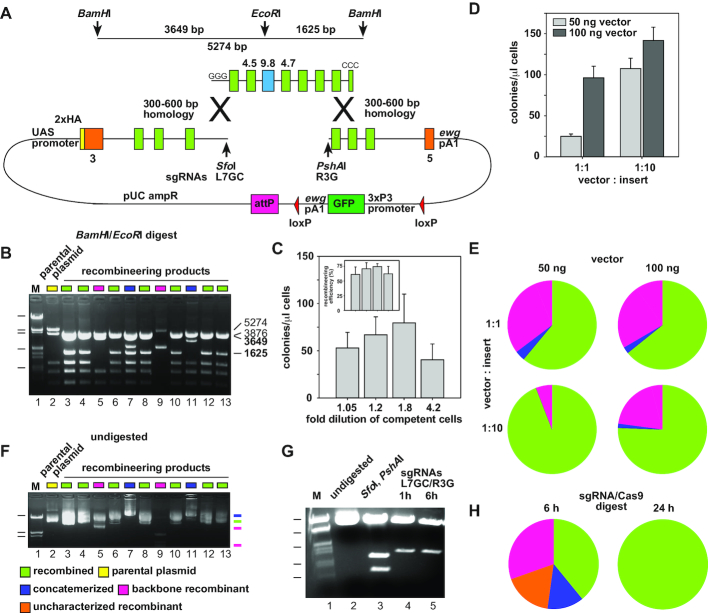
Figure 3.
Plasmid based gap repair recombineering for modification of large plasmids. (A) Schematic for λ Red protein mediated gap-repair recombineering of the pUC 3GLA HAi Dscam 3–5 plasmid linearized with SfoI and PshAI with a modified insert containing homologous sequences of the beginning and the end of the insert and vector. GGG at the beginning and CCC at the end of the insert indicate the genomic sequence of the SmaI sites used for excission from the pOT2 vector (not shown). On top, fragment sizes for fingerprinting the parental and recombineered constructs with BamHI and EcoRI are indicated. Homologous regions for recombination are indicated by crosses. (B) Agarose gel of representative recombinant plasmids fingerprinted by BamHI/EcoRI restriction digests. Correct recombinants (green squares) are identified by 3649 and 1625 bp fragments originating from the 5274 bp fragment in the parental vector due to the additional EcoRI site introduced by exon 9.8. The parental plasmid is indicated by yellow, backbone recombinants by pink, uncharacterized recombinants by orange, and concatemerized plasmids by blue squares. Size markers are EcoRI/HinDIII digested λ DNA of 20, 5.2, 3.5, 1.9 and 0.8 kb. (C) Effect of competent cell concentration on transformation and recombineering efficiency. Transformation efficiency is shown as mean with standard error for the number of colonies obtained per microliter of competent cells (normalized to a transformation efficiency of 106 transformants/μg of a 3 kb plasmid) from at least four experiments. Dilutions of the starting 25 μl of competent cells with 1.25 μl (1.05-fold dilution), 5 μl (1.2-fold), 20 μl (1.8-fold) and 80 μl (4.2-fold) are shown at the bottom. The insert shows the recombineering efficiency as percentage of positive clones using 50 ng vector with a vector to insert ratio of 1:10. (D) Effect of vector concentration and vector to insert ratio on transformation efficiency. Transformation efficiency is shown as mean with standard error for the number of colonies obtained per microliter of competent cells (normalized to a transformation efficiency of 106 transformants/μg of a 3 kb plasmid) for 50 ng (light gray) or 100 ng of vector (dark gray) and a vector to insert ratio of 1:1 or 1:10 from at least eight experiments each and a total of 372 clones. (E) Effect of vector concentration and vector to insert ratio on recombineering accuracy and efficiency. Results are shown as pie charts from at least four independent experiments analyzing a total of at least 50 clones each for 50 ng (left) or 100 ng vector (right) and a vector to insert ratio of 1:1 (top) or 1:10 (bottom). (F) Agarose gel of undigested plasmids shown in (B). Colored lines on the right side indicate positions of supercoiled plasmids. Size markers are EcoRI/HinDIII digested λ DNA of 20, 5.2 and 3.5 kb. (G) Agarose gel showing Cas9 mediated cleavage of the Dscam 3–5 plasmid with sgRNAs L7GC and R3G. (H) Extended digestion with sgRNA/Cas9 is required for full plasmid cleavage. Results are shown as pie charts from two independent experiments with 6h and 24 h digestion time with sgRNAs L7GC and R3G using 50 ng vector and a vector to insert ration of 1:10 for transformation.