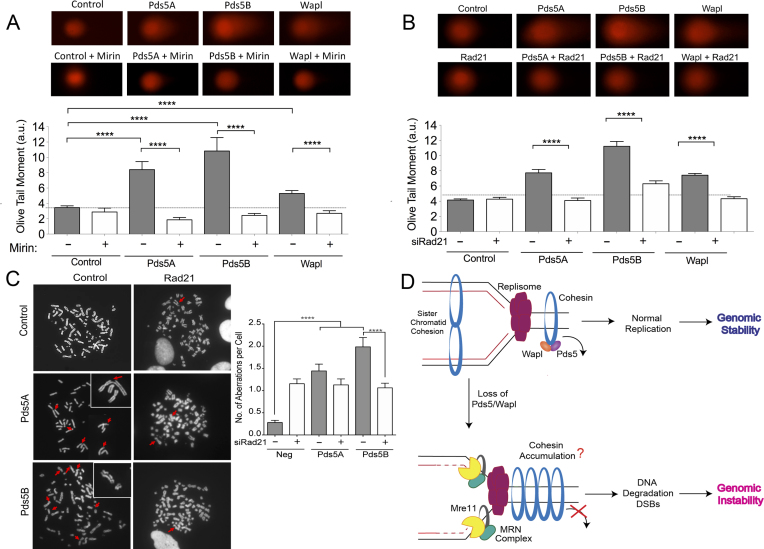
Figure 6.
Loss of Pds5 leads to increased DSBs and chromosomal aberrations. (A) Neutral Comet assay monitoring DSB formation in in Pds5A, Pds5B or Wapl siRNA depleted U-2 OS cells treated with 50 μM Mirin or left untreated. Representative images of comets of Pds5A, Pds5B, Rad21 or Wapl siRNA depleted cells with or without 50 μM Mirin (top). Out of three repeats; n ≥ 100 comets scored for each data set. Data are represented as mean ± SEM. Statistics: Mann–Whitney; ****P < 0.0001 (bottom). (B) Neutral Comet assay monitoring DSB formation in Pds5A, Pds5B, Wapl, Pds5A/Rad21, Pds5B/Rad21 and Wapl/Rad21 siRNA depleted U-2 OS cells. Representative images of comets of Pds5A, Pds5B, Rad21, Wapl, Pds5A/Rad21, Pds5B/Rad21 and Wapl/Rad21 siRNA depleted cells (top). Out of three repeats; n ≥ 100 comets scored for each data set. Data are represented as mean ± SEM Statistics: Mann–Whitney; ****P < 0.0001 (bottom). (C) Accumulation of DSB as measured by chromosome spread. Representative images of metaphases of Pds5A, Pds5B, Rad21, Pds5A/Rad21 and Pds5B/Rad21 siRNA depleted cells (left). Red arrows point to DSBs. Bar graph, distribution of number of chromosomal abnormalities per metaphase. At least 50 metaphases were counted from three independent experiments. Mean shown, n = 3. Statistics: unpaired t-test; ****P < 0.0001. (D) Proposed model. Cohesin is recruited to origins in early S-Phase. Pds5/Wapl unload cohesin ahead of the fork to allow fork passage. Cohesin is promptly re-assembled behind the replication fork to achieve sister chromatid cohesion. Whether the two sister chromatids are embraced by two interacting cohesin complexes, as shown in the figure, or by a single-complex is still controversial in the field (84–87). In the absence of Pds5 or Wapl, there is a reduction in cohesin loading behind the forks suggesting that there might be a consequent accumulation of cohesin rings ahead of replication forks. This aberrant re-distribution of the cohesin rings might cause MRE11-dependent degradation of the newly synthesized DNA strands, leading tract shortening, DSB accumulation and chromosomal instability. Tract shortening could also be due to an inhibitory effect of MRE11 on fork movement independent of fork degradation as discussed in the main text.