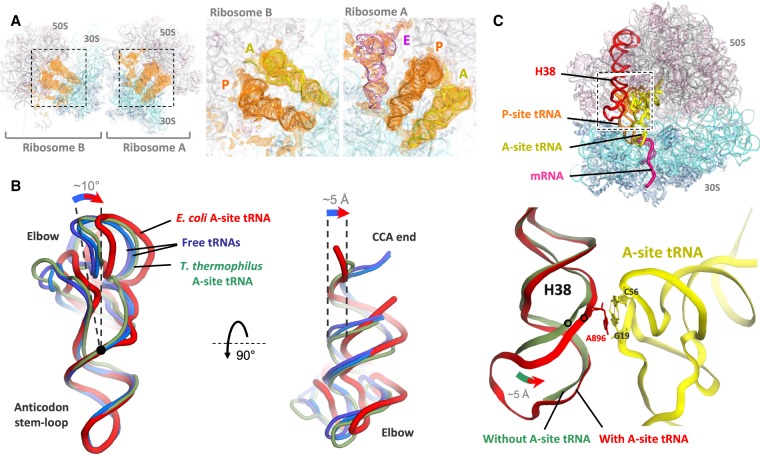
FIGURE 5.
tRNA structures in the hairpin complex. (A) Electron density peaks (orange) for tRNAs in mFo–DFc tRNA + mRNA omit maps for ribosomes A and B, contoured at 2σ and blurred by 200 Å2. The insets show the map overlaid on the tRNA models. (B) Cartoon representation of the A-site tRNA (red) viewed from the subunit interface (left) or from the large subunit (right). The A-site tRNA in the E. coli ribosome (red, this study) is bent compared to unbound tRNA (light and dark blue, PDB 1EHZ and 4TRA, respectively) (Westhof et al. 1988; Shi and Moore 2000). The nonbent A-site tRNA in the T. thermophilus ribosome (green, PDB 4V5D) (Voorhees et al. 2009) is also shown for comparison. Position of the 26:44 base pair is indicated by a black dot. All tRNA structures are aligned by their anticodon stem–loop. (C) In the presence of A-site tRNA (yellow), the tip of helix H38 of 23S rRNA (red) interacts with the tRNA elbow, moving by up to 5 Å toward it compared to when the A-site is vacant (faint green, PDB 4YBB) (Noeske et al. 2015). The stacking of A896 base of H38 on the G19:C56 tertiary base pair of the tRNA is shown. Note that the terminal loop of H38 was not modeled in the vacant complex.