Figure 6.
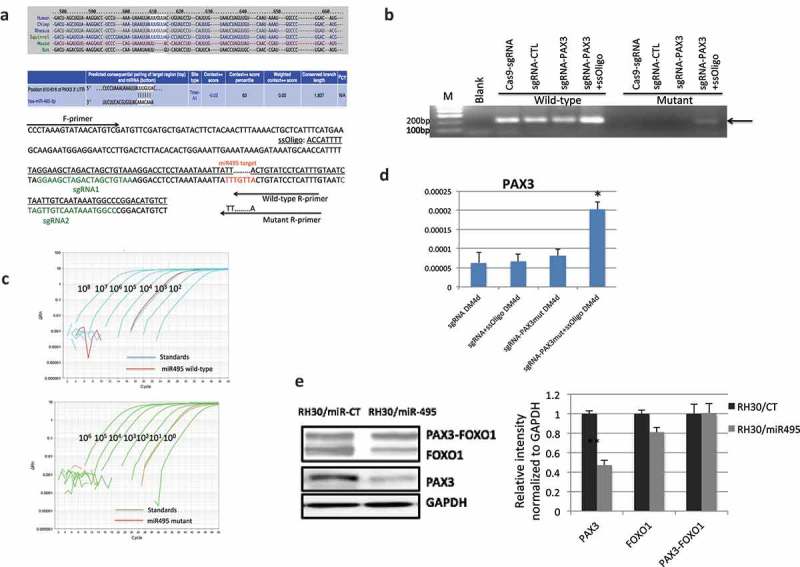
CRISPR/Cas9 mediated genome editing at miR-495 seed sequence. (a) The location and sequence of the sgRNAs, and the template ssOligo for CRISPR/Cas9 editing. miRNA-495 seed sequence and primers specific to the wild-type and mutated alleles were also plotted. (b) Allele-specific primers picked up the mutant form only when both sgRNAs and the ssOligo template were included. (c) Relative number of the mutated alleles to the wild-type alleles after CRISPR/Cas9 mediated genome editing. Quantitative PCR revealed that the relative number of mutated alleles to the wild-type alleles is close to 1/1000. Purified PCR products were serial diluted, and used as templates to construct the standard curve. (d) PAX3 RNA was increased during human MSC muscle differentiation when miR-495 seed sequence was changed to the corresponding sequence of mice. (e) miR-495 suppressed endogneosu PAX3, but not the fusion. RH30 cells were transfected with miR-495 mimics or the control miR-CT. Western blot analysis were performed using FOXO1, PAX3, and GAPDH antibodies (left). Quantification of protein signal intensity was conducted using densitometry, and plotted on the right. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
