Figure 2.
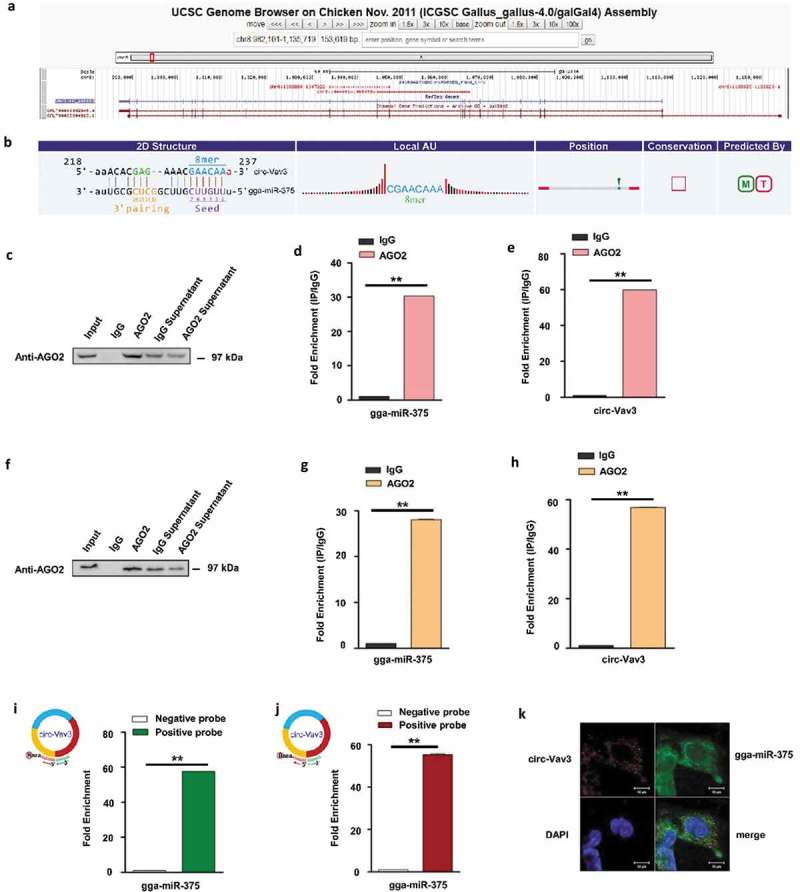
Circ-Vav3 sponges with gga-miR-375. (a) The Ensembl screen shot of the circ-Vav3 locus associated with the Vav3 gene. (b) Prediction and sequence analysis of gga-miRNA-375 response elements in circ-Vav3. The 2D structure shows the gga-miR-375 response element sequence and the target gga-miR-375 seed type (8-mer) and 3ʹ pairing sequence (nucleotides 13–16). The precise base positions are shown in the alignments in the upper left and right corners. (c and f) RIP assay shows that the miRNAs were successfully pulled-down using an AGO2 antibody. IgG was used as the negative control. (d, e, g and h) Fold enrichment of gga-miR-375 and circ-Vav3, respectively, quantified by RT-qPCR after RNA immunoprecipitation. GAPDH was the reference gene of circ-Vav3, U6 (figure d) and 5S (figure g) were used as the reference gene of gga-miR-375 (** represent P < 0.01). (i and j) Fold enrichment of gga-miR-375 quantified by RT-qPCR after circ-Vav3 pull-down. U6 (figure i) and 5S (figure j) were used as the reference gene of gga-miR-375 (** represent P < 0.01). (k) RNA in situ hybridization reveals the co-localization of circ-Vav3 and gga-miR-375 in the cytoplasm of DF-1 cells after the co-incubation of biotin-labeled circ-Vav3 and digoxin-labeled gga-miR-375 probes. Nuclei were stained with DAPI. Scale bar, 10 μm. The data represent the mean±s.e.m. of three independent experiments.
