Figure 2.
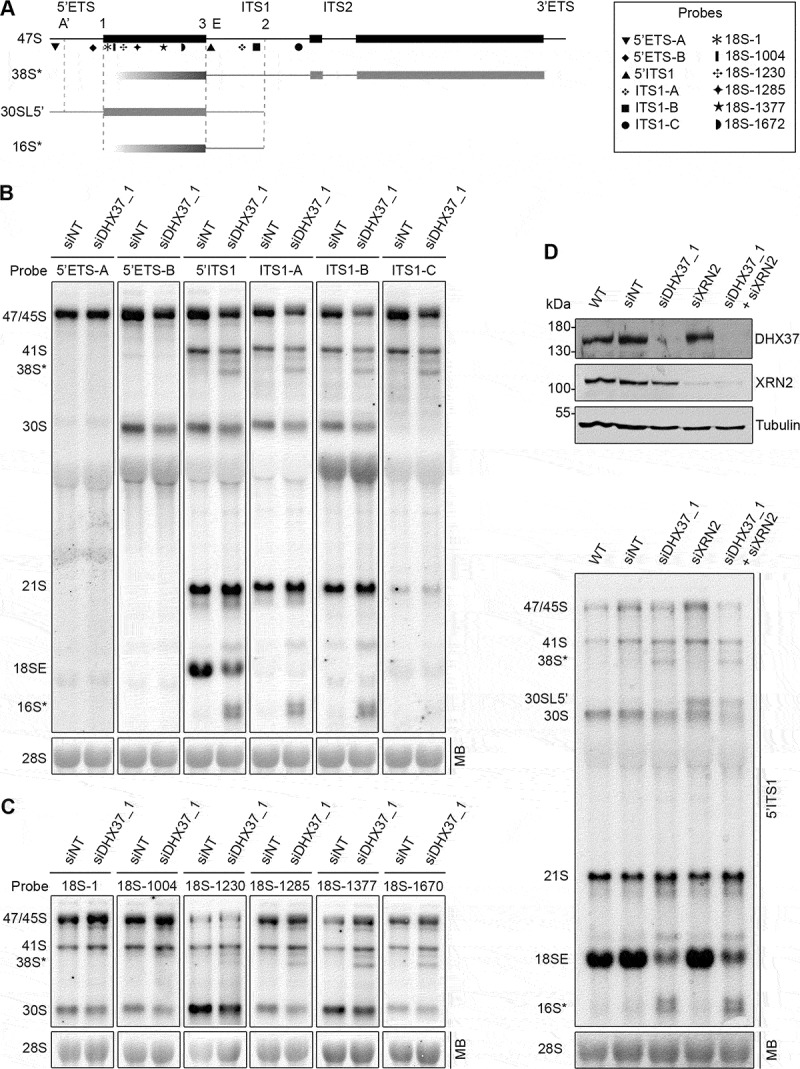
Mapping of the aberrant 16S* and 38S* pre-rRNA intermediates that accumulate upon depletion of DHX37. (a) Schematic view of the 47S pre-rRNAs (black) and the aberrant 38S*, 16S* and 30SL5ʹ pre-rRNA species not normally detected in human cells (grey). Mature rRNA regions are shown as rectangles, and internal (ITS) and external transcribed spacer (ETS) regions are represented by lines. Cleavage sites are named above the 47S pre-rRNA and the hybridization position of probes used for northern blotting are indicated. (b,c) RNA extracted from HeLa cells transfected with non-target siRNAs (siNT) or siRNAs targeting DHX37 (siDHX37_1) was separated by denaturing agarose gel electrophoresis, transferred to a nylon membrane and the mature 28S rRNA was detected by methylene blue staining (MB). Northern blotting was performed using probes hybridizing to different positions within the 5ʹ ETS and ITS1 (b) or the 18S rRNA (c) and pre-rRNAs were visualized using a phosphorimager. (d) RNA and proteins were extracted from wildtype (WT) HeLa cells or cells that had been transfected with non-target siRNAs (siNT), or siRNAs targeting DHX37 (siDHX37_1) or XRN2 (siXRN2). Proteins were analyzed by western blotting using the antibodies indicated to the right (upper panel) and pre-RNAs were detected by northern blotting using a probe hybridizing to the 5' end of ITS1.
