Figure 6.
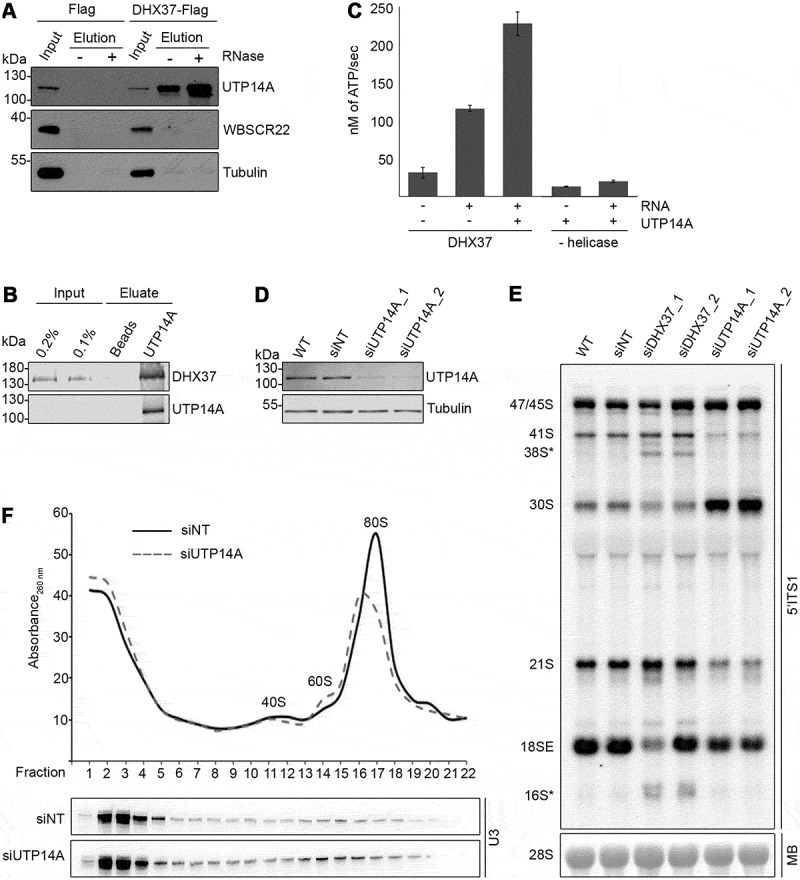
UTP14A interacts with DHX37 and stimulates its ATPase activity in vitro. (a) Cell extracts prepared from HEK293 cells expressing either DHX37-Flag or the Flag tag were used for immunoprecipitation experiments in the presence (+) or absence (-) of RNase. Inputs (1%) and eluates were separated by SDS-PAGE and analyzed by western blotting using antibodies against DHX37, UTP14A, WBSCR22 and tubulin. (b) Recombinant DHX37-His6 was incubated with either IgG sepharose (beads) or with IgG sepharose on which ZZ-UTP14A-His10 had been immobilized. After washing steps, proteins were eluted, and inputs and eluates were separated by SDS-PAGE then analyzed by western blotting using antibodies against DHX37 and UTP14A. (c) The amount of ATP hydrolyzed by recombinant DHX37 in the presence (+) or absence (-) of RNA and the presence (+) or absence (-) of UTP14A was determined using an in vitro NADH-coupled ATPase assay. Experiments were performed in triplicate and error bars represent mean ± standard deviation. (d) HeLa cells were left untransfected (WT), or were transfected with non-target siRNA (siNT) or siRNAs targeting UTP14A (siUTP14A_1 and siUTP14A_2). After 72 h, cells were harvested, and proteins were analyzed by western blotting using antibodies against UTP14A (upper panel) and tubulin (lower panel). (e) Total RNA was extracted from wild-type (WT) HeLa cells or HeLa cells transfected with siRNAs as in (D). RNAs were separated by denaturing agarose gel electrophoresis, transferred to a nylon membrane and the mature 28S rRNA was visualized by methylene blue staining (MB). Northern blotting with a probe hybridizing to the 5ʹ end of ITS1 (5ʹ ITS1) was used to detect precursors of the 18S RNA, which were visualized using a phosphorimager. (f) Whole cell extracts prepared from HEK293 cells treated with control siRNAs (siNT) or siRNAs targeting UTP14A (siUTP14A_1) were separated by sucrose density gradient centrifugation. The absorbance of each faction at 260 nm was determined and used to generate a profile on which the peaks corresponding to ribosomal subunits (40S and 60S) and 80S monosomes are indicated. RNA extracted from the gradient fractions was separated by denaturing PAGE, transferred to a nylon membrane and northern blotting was performed using a [32P]-labelled probe hybridizing to the U3 snoRNA. Replica experiments have been performed and representative data is shown in this figure.
