Figure 6.
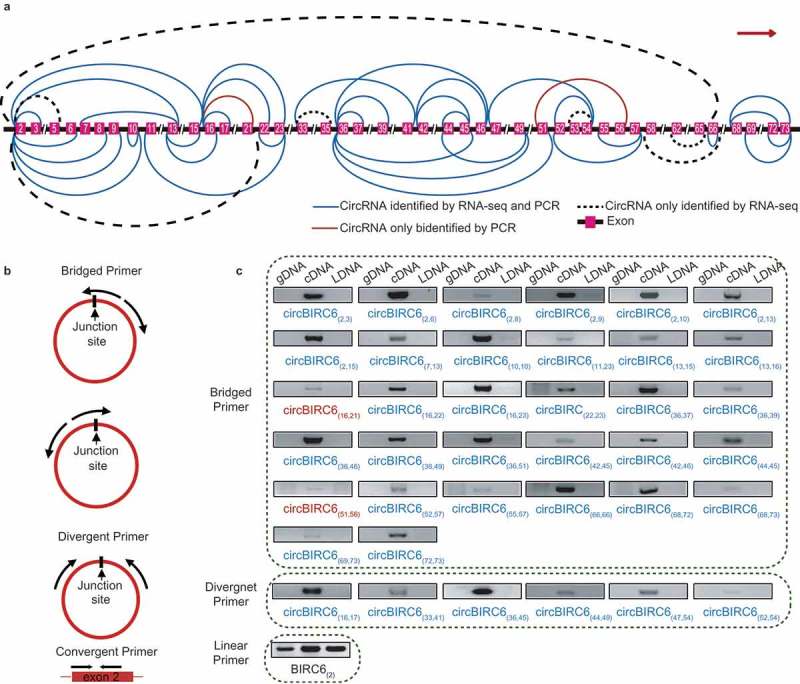
Forty-five isogenous circRNAs derived from the BIRC6 gene. (a) Schematic diagram showing the 45 isogenous circRNAs derived from BIRC6 gene. Forty-three isogenous circRNAs were identified in the RNA-seq analysis. Among them, 36 circRNAs were validated by PCR (described as ‘CircRNAs identified by RNA-seq and PCR’); Seven circRNAs failed to be validated by PCR (described as ‘CircRNAs only identified by RNA-seq’). Two circRNAs were only verified by PCR (described as “CircRNAs only identified by PCR). The colored lines indicate the start and end positions of the circRNAs in the gene. (b) Schematic diagram showing the bridged and divergent primer amplification strategies. The bridged primer is shown to span the exon junctions, so as to only amplify and verify the circRNAs using cDNA templates. The divergent primers are shown to amplify the back-splicing exon junction regions of circRNA; the convergent primers are shown to amplify the exon regions as controls. (c) PCR verification of 38 BIRC6 isogenous circRNAs using bridged and divergent primer amplification strategies. Thirty-two circRNAs were validated by the ‘bridged primer amplification strategy’ using cDNA templates; as controls, the PCR products were not amplified from gDNA and LDNA (the complementary DNA reverse transcribed using oligo dT primers). Among them, 30 circRNAs were also identified by RNA-seq analysis; two circRNAs were novel. Six circRNAs were validated by the ‘divergent primer amplification strategy’. Convergent primers were used to amplify exon 2 as controls.
