ABSTRACT
The exosome functions to regulate the cellular transcriptome through RNA biogenesis, surveillance, and decay. Mutations in Dis3, a catalytic subunit of the RNA exosome with separable endonuclease and exonuclease activities, are linked to multiple myeloma. Here we report that a cancer-associated DIS3 allele, dis3E729K, provides evidence for DIS3 functioning in mitotic fidelity in yeast. This dis3E729K allele does not induce defects in 7S→5.8S rRNA processing, although it elicits a requirement for P-body function. While it does not significantly influence cell cycle progression alone, the allele reduces the efficiency of cell cycle arrest in strains with defects in kinetochore assembly. Finally, point mutations in the exonuclease domains of yeast Dis3 elicit genome instability phenotypes; however, these DIS3 mutations do not increase DNA damage or RNA processing defects that lead to the accumulation of polyadenylated RNA in the nucleus. These data suggest that specific DIS3 activities support mitotic fidelity in yeast.
KEYWORDS: DIS3, exosome, genome instability, Saccharomyces cerevisiae, mitotic spindle, SGA
Introduction
Dis3 is one of two catalytic components, along with Rrp6, of the eukaryotic RNA exosome complex. Dis3 is a 3ʹ-5ʹ exonuclease and endonuclease that is conserved from bacteria to humans, and participates in the processing and degradation of many RNA species [1] . DIS3 mutants suffer from impaired RNA processing, defective microtubules, growth retardation, and temperature sensitivity [2–4]. Dis3 also participates in kinetochore assembly in Schizosaccharomyces pombe by contributing to pericentromeric chromatin silencing [4]. Most recently, studies in Drosophila melanogaster and Caenorhabditis elegans revealed that Dis3 is a target of CDK1 phosphorylation, and this phosphorylation reduces the Dis3 exonuclease function in the G2 phase of the cell cycle [5]. Having multiple enzymatic activities, contributing to almost all aspects of RNA metabolism, and showing pleiotropic phenotypes upon mutation has made the mechanisms by which Dis3 contributes to each phenotype difficult to determine.
Delineating the impact of DIS3 perturbation has become of medical importance over the past decade as DIS3 mutations have been identified in roughly 11% of multiple myeloma (MM) patients, particularly within the exonuclease domain [6]. MM is a genetically heterogeneous plasma cell neoplasm, responsible for 10–15% of all blood malignancies, and is characterized by activation of MYC, KRAS, NRAS, and FGFR3 in addition to a host of recurrent aneuploidies, including loss of 13q14 and 17p13 [6–8]. Reduction-of-function mutations in DIS3 seem to arise early in tumorigenesis, implicating DIS3 as a potential tumor-suppressor gene [7].
Genome instability is a hallmark of many cancers, which provides cancer cells with enhanced evolutionary capacity by increasing the potential for sequence and karyotypic changes [9]. Genome instability can be subdivided into microsatellite instability (MIN) and chromosome instability (CIN), which induce increases in mutation rate and the rate of aneuploidy, respectively [10]. CIN is generally characterized by whole chromosome gain or loss, recurrent breakage events and/or gross chromosomal rearrangements [11]. Due to the inherent molecular complexity of these events, the cellular circuits that sustain these phenotypes remain to be fully characterized. Large screens using model organisms such as Saccharomyces cerevisiae have allowed for the comprehensive identification of genes and pathways that when disrupted cause CIN [12,13]. Having identified a temperature sensitive (ts) allele of DIS3 among the novel hits, the challenge is to now understand the mechanisms by which Dis3 and the many other identified factors contribute to the maintenance of genome stability.
Towards this goal, we have phenotypically characterized a yeast strain harbouring a Dis3 mutation (E729K) that is orthologous to a human mutation (E665K) first identified in a myeloma sequencing study [7]. Analysis by synthetic genetic array (SGA) identified synthetic growth defects between this DIS3 mutant and spindle assembly checkpoint proteins and kinetochore components. The dis3E729K mutant has been studied alongside control strains that are temperature sensitive or have endonuclease or exonuclease deficiency; these strains have allowed us to specifically link Dis3 exonuclease domain function to chromosome stability, with exonuclease mutants exhibiting reduced fitness and increased CIN. Together these data link genome maintenance to the DIS3 exonuclease domain through a mechanism likely involving the mitotic chromosome segregation apparatus.
Results and discussion
Characterization of DIS3 alleles
Previously, a DIS3-temperature sensitive (ts) allele, dis3-ts, was found to exhibit a weak chromosome transmission fidelity (ctf) phenotype [14,15]. We sequenced the dis3-ts allele to investigate this phenotype with respect to Dis3 activity but found that dis3-ts carries 10 non-synonymous variants throughout the length of the gene (Figure 1(a)). As a result, it is difficult to link the ctf phenotype associated with this allele to any particular Dis3 domain or activity, or assess the relevance of these findings to the phenotypes of cancer cells carrying DIS3 mutations. Thus, we engineered a disease-relevant single point mutation into budding yeast DIS3 to investigate the influence of this mutation, in comparison to dis3-ts.
Figure 1.
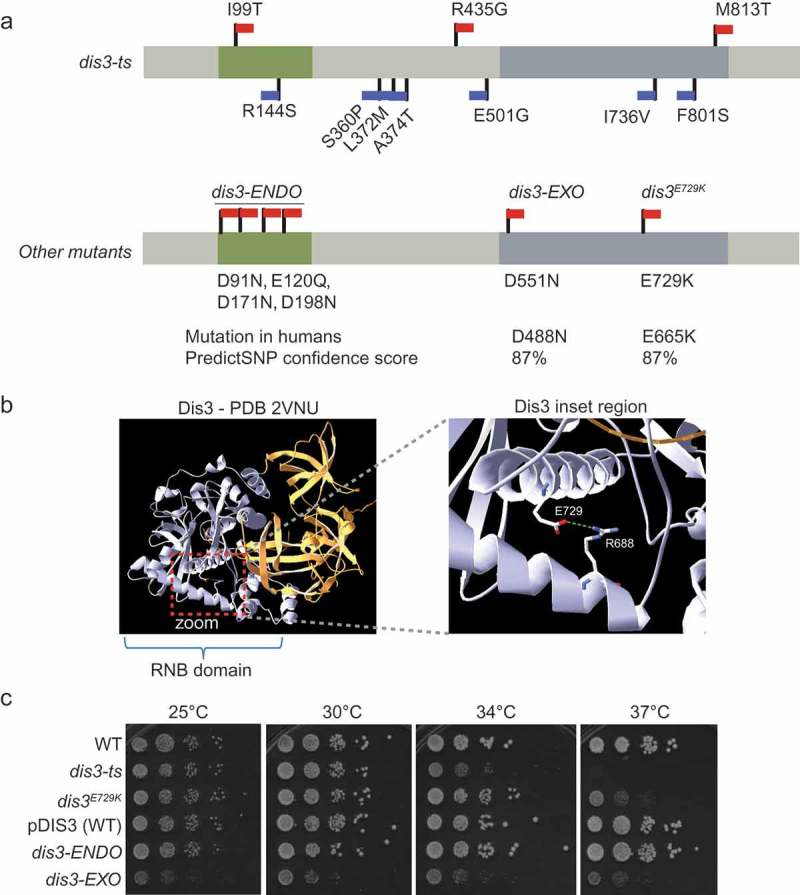
Characterization of DIS3 separation-of-function, cancer-associated and ts-alleles. (a) Structure of major DIS3 domains, with the non-synonymous mutations in dis3-ENDO, dis3-EXO, dis3E729K, and dis3-ts indicated. Red flags indicate mutations predicted to be deleterious by PredictSNP; blue flags indicate predicted neutral mutations [16]. (b) Structure of Dis3 protein highlighting the E729 residue. The RNB domain is coloured grey, while the N-terminus is coloured gold. A zoomed inset of the helices containing E729 and its hydrogen bonding partner R668 are shown on the right (a green dashed line shows the predicted H-bond) [18]. (c) Ten-fold serial dilution spots of wildtype controls versus DIS3 mutants. Plates were incubated at the indicated temperature for 2 days prior to scanning.
We chose a poorly-characterized multiple myeloma-associated point mutation, human dis3E665K, that is located within the exonuclease domain of Dis3. In yeast, dis3E729K is orthologous to the human E665K mutation and likely to be deleterious according to PredictSNP [16,17]. Analysis of a Dis3 crystal structure shows that the E729 sidechain can make a hydrogen bond contact with the guanidino moiety of R688 (Figure 1(b)) [18]. The E729K substitution would disrupt this bond, and change an acidic residue for a basic residue, which is likely to distort and/or destabilize this region of the RNB domain. In addition, we employed two control strains bearing mutations that inactivate either the endonuclease (dis3-ENDO; D91N, E120Q, D171N, and D198N), or the exonuclease (dis3-EXO; D551N) activity of Dis3 [19]. These strains carry the mutant DIS3 allele on a plasmid with the genomic copy of DIS3 deleted; for this reason, a plasmid-borne version of wildtype DIS3 is used as an additional control (‘pDIS3-WT’). As expected, and in agreement with previous studies [2], dis3-EXO suffers from a growth defect at all tested temperatures, while dis3-ENDO appears to maintain near wildtype growth under these conditions (Figure 1(c)). Comparing the growth of these strains to dis3-ts and dis3E729K revealed that dis3E729K exhibits considerable temperature sensitivity (Figure 1(c)). It is notable that dis3-EXO also corresponds to a recurrent mutation observed in multiple myeloma [17].
Genomic profiling of the cancer-associated dis3e729k indicates mitotic defects
We wished to determine if this cancer-associated DIS3 point mutation (i.e. dis3E729K) induced cellular defects similar to those previously reported for dis3-ts. An unbiased way to identify cellular pathways depending on Dis3 function is through genetic interaction profiling by synthetic genetic array (SGA) [20]. We predicted that genes in pathways that are buffering specific defects in DIS3 mutants would appear as negative interactions in the SGA data. We profiled both the dis3-ts and dis3E729K alleles, as this would facilitate a direct comparison with previously reported DIS3 SGA datasets (collected from a different DIS3 allele). As well, we anticipated that the comparison would allow us to determine if the difference in the fitness of these mutants (i.e. temperature sensitivity at 37°C) represents a difference in the cellular effects of dis3-ts versus dis3E729K, or simply difference in severity of the same phenotypes.
We screened these two alleles against the yeast deletion collection comprised of non-essential gene knockouts, and collections of DAmP (Decreased Abundance by mRNA Perturbation) and ts-alleles representing >80% of all essential genes [21–23]. In total, dis3-ts and dis3E729K had candidate negative genetic interactions surpassing our cut-off with 94 and 99 genes, respectively (Figure 2(a), Table S1). We compared these gene lists to that published as part of a large scale study for dis3-1 [20]. At the level of individual genes, there is little overlap in the genes identified as negative genetic interactors of DIS3 between the tested alleles. However, at the level gene ontology (GO) cellular components, all three DIS3 alleles elicit general genetic interactions with nuclear components, while dis3-ts and dis3-1 specifically enrich for components of the exosome and those involved in ribosome biogenesis, a known function of Dis3 [24] (Figure 2(b) and Table S2). In contrast, dis3E729K and dis3-ts enrich specifically for components of the spindle pole body/microtubule organizing centre. The shared GO terms found between dis3-ts and the other two alleles suggests that the numerous point mutations in dis3-ts may be impact multiple Dis3 functions, while dis3-1 and dis3E729K have interactions that are restricted to more discrete activities. We validated synthetic sick genetic interactions between dis3E729K and several SGA hits with diverse functions, including mutants involved in ribosome biogenesis (nop2-5, bms1-1), and mitotic mutants influencing the kinetochore (e.g. dam1-1, spc24 4–2, mif2-3), the Anaphase Promoting Complex (apc2-1), or the spindle pole body (cdc31-2) with spot dilution plating assays (Figure 2(c) and noted in Table S1). Together, the high-resolution phenotypes provided by these SGA screens suggests allele specificity amongst DIS3 mutants and that dis3E729K is genetically linked to the mitotic chromosome segregation apparatus.
Figure 2.
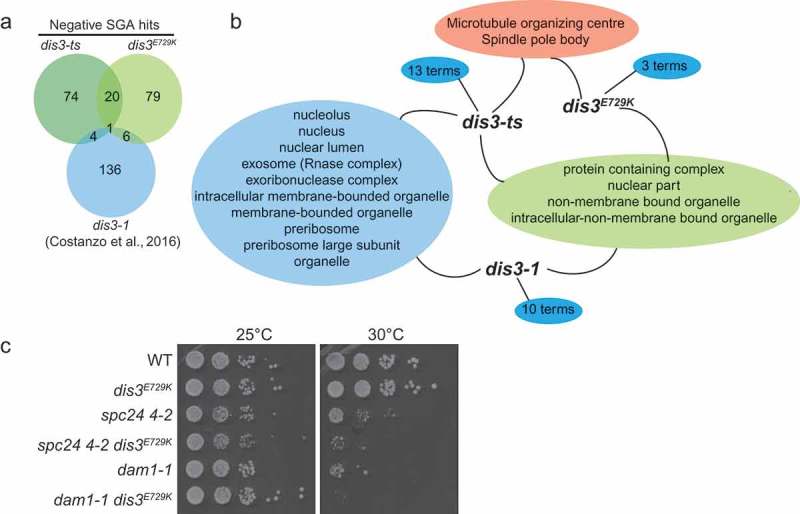
Genomic profiling of dis3E729K and dis3-ts indicates potential mitotic defects. (a) Overall results showing negative genetic interactions from two screens in this study, and the published dis3-1 SGA screen [20]. (b) Analysis of gene ontology terms among three gene lists reveals specialization of DIS3 mutant allele phenotypes. (c) Spot-dilution assay validations of negative genetic interactions between spc24 or dam1 mutant alleles with dis3E729K.
DIS3 mutations induce distinct RNA processing defects
Given that the SGA results for dis3E729K revealed different GO term enrichments compared with dis3-1 or dis3-ts, we wished to assess if this allele would exhibit RNA processing or degradation defects that have been previously reported in DIS3 loss-of-function mutants. We performed northern blotting to address 5.8S RNA maturation in our panel of DIS3 mutants. As expected based on the literature and SGA screen results, we observed substantial accumulation of 5.8S processing intermediates and precursors in dis3-ts and dis3-EXO at 37°C. In contrast we observed essentially wildtype 5.8S processing in dis3E729K and dis3-ENDO (Figure 3(a)). This is consistent with the SGA results, as rRNA processing related terms were abundant in the dis3-ts (Table S2) and dis3-1 datasets, yet were not enriched in the dis3E729K data. Notably, dis3E729K has strong growth defects at 37°C, suggesting that these growth defects arise for reasons other than changes in 5.8S rRNA processing.
Figure 3.
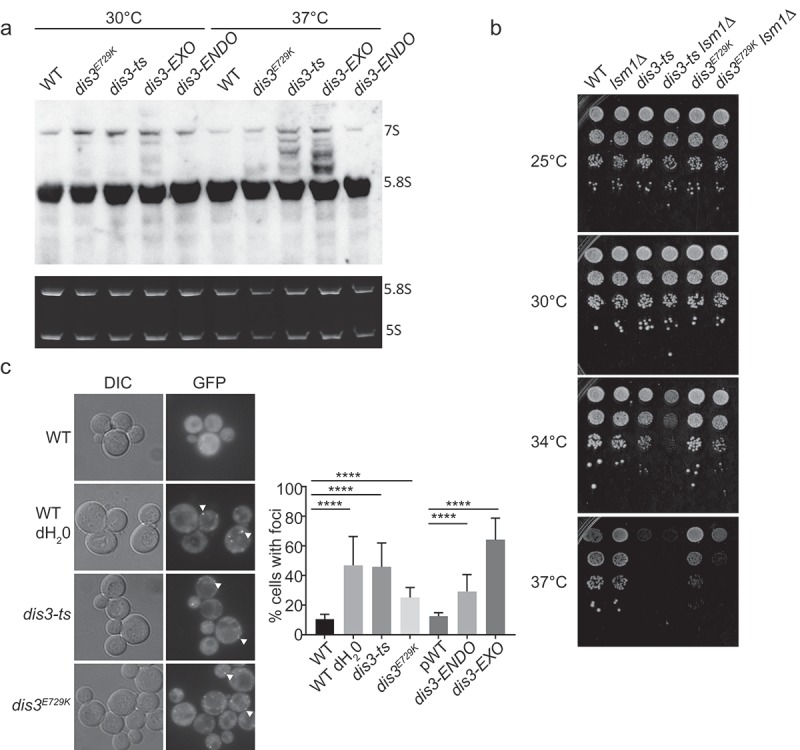
RNA processing phenotypes of DIS3 alleles. (a) Northern blot of 5.8S rRNA maturation products in early-log phase cultures. The top panel is a 15 second exposure after hybridization with a 5.8S rRNA-specific probe, and bottom panel is an ethidium bromide stained gel demonstrating RNA loading. (b) Spot dilution assays showing genetic interaction of dis3-ts and dis3E729K with loss of the P-body component Lsm1. (c) Representative images (left) and quantification (right) of Lsm1-GFP labelled P-body formation in DIS3 mutant alleles. Arrows indicate representative foci. **** indicates a p < 0.0001 via a Fisher exact test with Holm-Bonferroni correction.
Since 5.8S is just one of many transcripts engaged by Dis3, we sought a more general assay to measure cellular stress in these mutant strains. Cellular stress is known to induce requirements for parallel RNA degradation activities localized in cytoplasmic Processing-bodies (P-bodies) [25,26]. When we deleted a P-body component and mRNA 5ʹ decapping factor, LSM1, we found that the dis3-ts and dis3E729K fitness defects were greatly enhanced at 37°C, with dis3-ts also showing a clear growth defect at 34°C (Figure 3(b)). In support of ongoing cellular stress, all DIS3 mutants were found to exhibit enhanced P-body formation based on Lsm1-GFP localization in the absence of exogenous stress (Figure 3(c)), but dis3E729K had the lowest levels, more similar to that of the dis3-ENDO allele. These data further support the idea that dis3E729K has a weaker general RNA processing defect compared to other exonuclease mutants and/or may be impacting a discrete function of Dis3. Interestingly, loss of LSM1 has also been linked to genome instability through at least two mechanisms: one study suggested the P-bodies regulate the levels of histone mRNA during replication [27], while another study showed that P-bodies regulates the transcriptional repressor YOX1, leading to oxidative stress and DNA damage in lsm1Δ [28]. Whether these phenotypes are relevant to the observed genetic interactions with DIS3 mutants are unknown, although the DNA damage phenotypes in lsm1Δ strains are not seen in our DIS3 mutants (see below).
Finally, since temperature sensitive dis3 alleles, and other exosome mutants, have been reported to accumulate poly(A)-RNA in the nucleolus at the non-permissive temperature [29], we asked whether this phenotype is present in dis3E729K . Fluorescence in situ hybridization (FISH) revealed that poly(A)-RNA accumulation after a 37°C temperature shift was specific to dis3-ts and did not occur appreciably in dis3-ENDO, dis3-EXO, or dis3E729K (Figure S1). Similarly, probes directed against the 5.8S rRNA and second internal transcribed spacer sequence (ITS2) region of the rRNA precursor transcript showed defects in rRNA processing in dis3-ts through a decrease in the nucleolar signals for both probes, which is not observed in the other mutants. Thus, while dis3-EXO and dis3-ts exhibited defects in 5.8S rRNA processing (Figure 3(a)), these changes were not accompanied by poly(A)-RNA accumulation or strong alterations in nucleolar staining with 5.8S rRNA or ITS2 probes in dis3-EXO. These differences again suggest that Dis3 functions within the cell are differentially perturbed at 37°C by the mutations present in the tested alleles.
The dis3e729k allele alters cell cycle progression in the context of kinetochore deficiency
The observed phenotypic differences amongst these DIS3 alleles in the context of our SGA analysis indicates that cell cycle functions, particularly during anaphase, are involved in the maintenance of cellular fitness in dis3E729K and dis3-ts. In order to assess anaphase integrity in these mutants, we plated the DIS3 mutant strains on solid media containing microtubule poisons (benomyl and nocodazole). Both dis3-ts and dis3E729K showed resistance to benomyl (Figure 4(a)), and this was also evident on nocodazole plates for dis3-ts (Table 1). Previous work by Smith et al. demonstrated a benomyl and nocodazole sensitivity in mtr17-1 [3], another DIS3 temperature sensitivity allele, which is similar to the sensitivity seen in dis3-EXO at 30°C (Figure 4(a)). To further characterize this set of DIS3 alleles, we exposed strains to a range of stresses, including DNA damaging agents (MMS), disruptors of DNA replication (hydroxyurea), a nucleotide poison (5-fluorouracil), and a topoisomerase poison (camptothecin). A summary of these results is shown in Table 1. In general, DIS3 alleles had similar sensitivity to WT, or exhibited slight resistance to these stress treatments. Cumulatively, these data suggest that DNA damage sensitivity is not impacted by Dis3 mutations and confirms that mitotic function is likely dysregulated in DIS3 mutants, which results in varied sensitivities to microtubule poisons.
Figure 4.
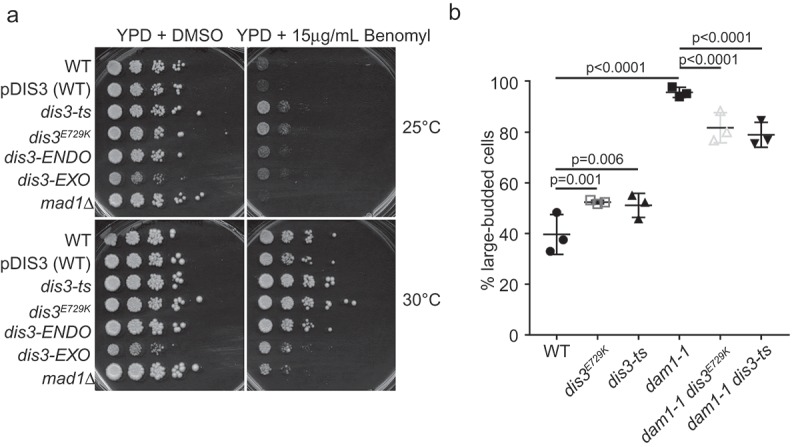
Mitosis-associated defects in DIS3 mutants. (a) Spot dilution assays of the indicated alleles on plates with microtubule depolymerizer benomyl. (b) Budding index measurements of the indicated mutants show suppression of dam1-1 G2/M arrest after a 3 hour temperature shift to 37°C by dis3E729K or dis3-ts. The Holm-Bonferroni corrected p-value determined by a Fisher Exact test is shown above each relevant comparison.
Table 1.
Growth behaviour of DIS3 mutant alleles under the indicated stress.
| Fitness change (relative to wildtype) |
||||||
|---|---|---|---|---|---|---|
| Temperatures tested | Condition (YPD +) | dis3-ts | dis3E729K | pDIS3-WT | dis3-Endo | dis3-Exo |
| 25°C, 30°C, 37°C | 5-FU 20 μM | 0 | 0 | 0 | 0 | 0 |
| 25°C, 30°C, 37°C | MMS 0.01% | 0 | 0 | 0 | 0 | 0 |
| 25°C, 30°C, 34°C, 37°C | HU 100 mM | 0 | 0 | 0 | 0 | 0 |
| 25°C, 30°C, 34°C, 37°C | Camptothecin 10 μg/ml | 0 | 0 | - | - | - |
| 25°C, 30°C, 34°C, 37°C | Nocodazole 4 μg/ml | + | 0 | 0 | + | + |
| 25°C, 30°C, 34°C, 37°C | Benomyl 15 μg/ml | + | + | 0 | + | + |
*(0 = no change, – = sensitive, + = resistant)
To begin to investigate the underlying cellular conditions that could be conferring the noted benomyl resistance, we first tested the budding index of dis3-ts and dis3E729K. These DIS3 mutants exhibited small but significant increases in the proportion of large-budded cells suggesting a G2/M delay (Figure 4(b)). We next combined the dis3-ts or dis3E729K alleles with the kinetochore mutant allele dam1-1. While dam1-1 alone exhibited an extreme increase of large budded cells compared to WT, indicative of a G2/M cell cycle arrest, the introduction of either the dis3E729K or dis3-ts allele significantly reduced the percentage of large budded cells (Figure 4(b)). This suggests that mutations in DIS3 alter aspects of mitotic arrest arising from kinetochore dysfunction, potentially explaining the observed changes in benomyl sensitivity among dis3 mutants, the dis3-ts ctf phenotype, and negative genetic interactions observed with alleles of DIS3 and the mitotic chromosome segregation apparatus (e.g. dam1-1 dis3E729K (Figure 2(c)). Alternatively, it is also possible that dis3-ts and dis3E729K may slow the cell cycle and influence benomyl resistance and mitotic fidelity, as it was recently shown that slowing the yeast cell cycle can improve mitotic fidelity by allowing time for normal corrective mechanisms to work [30].
Mutations in the DIS3 exonuclease domain cause genome instability, but not DNA damage
Previous work showed that dis3-ts exhibited a CIN phenotype using an artificial chromosome fragment loss reporter [13]. Our data suggests that dis3E729K may also induce CIN, through its influence on mitotic progression in the context of kinetochore dysfunction. To test CIN on endogenous chromosomal markers, and extend this to include the dis3E729K allele, we measured instability of chr III using the A-like faker (ALF) assay that measures loss of the native MAT locus [31,32]. We observed a significant increase in the frequency of ALF colonies for both dis3-ts and dis3E729K (Figure 5(a,b)). We observed no increase, relative to WT for the dis3-ENDO strain, although the ALF rates were higher than those of the pDIS3-WT. Remarkably, when we tested ALF in the dis3-EXO allele we saw a dramatic 68-fold increase in mating frequency relative to WT (Figure 5(b)). While it is unclear why CIN rates are this high in dis3-EXO, we note that this allele completely ablates the exonuclease activity and shows slow growth at all tested temperatures, whereas dis3-ts and dis3E729K only show strong growth defects at temperatures above 34°C.
Figure 5.
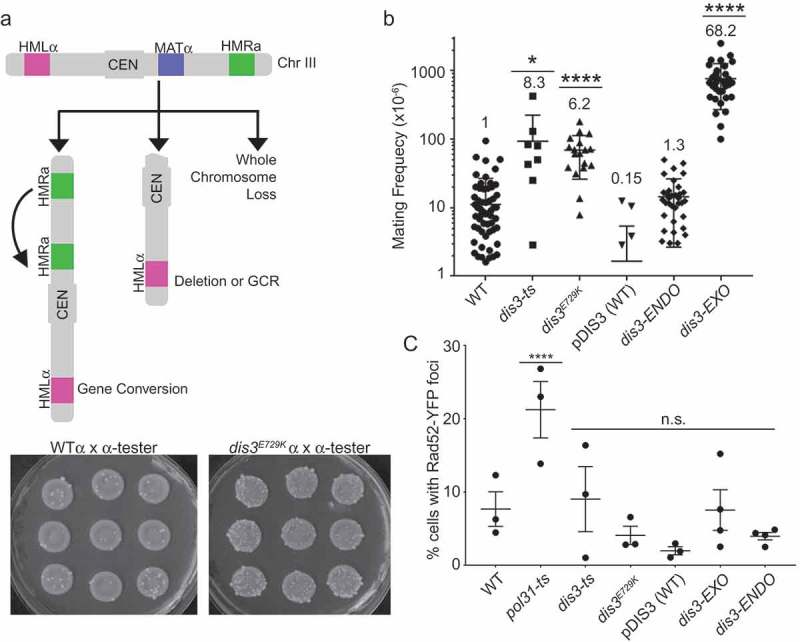
Mutations in the DIS3 exonuclease domain cause genome instability. (a) Schematic of ALF assay. Chromosome III has active (MAT) and silent mating loci (HMR, HML). Loss of MAT by gene conversion, deletion or whole chromosome loss leads to α-α mating. Sample images of nine independent mating rate tests in WT or dis3E729K are shown below. (b) Observed frequencies of mating in ALF assay for indicated strains. The fold increase over WT is shown above each dot plot. (c) Quantification of Rad52-YFP foci formation in DIS3 mutants. For B, data were compared with a Kruskal-Wallis Test. For C, Fisher’s Exact Test was used on raw count data. *p < 0.05, ****p < 0.0001.
ALF phenotypes can reflect multiple forms of CIN, including whole chromosome loss, local deletions or chromosome arm loss, and gene conversion [31,32]. One mechanism of CIN in RNA processing mutants that could lead to an ALF phenotype is the accumulation of DNA:RNA hybrids in genomic DNA called R-loops [33]. Indeed, previous work has found that the exosome accessory complex TRAMP, and the alternative catalytic subunit Rrp6 are involved in suppressing R-loop associated CIN [34–36]. We assessed R-loop levels by staining chromosome spreads from the strains with the S9.6 antibody, which recognizes DNA:RNA hybrids [37]. While this assay confirmed the previously reported R-loop increase in dis3-ts, we did not find evidence for R-loops accumulating in dis3E729K. In addition, S9.6 staining did not indicate excessive R-loops accumulating in dis3-EXO, although we did observe a small but significant increase in this strain relative to the plasmid WT control (Figure S2). Thus, staining with S9.6 did not correlate with the CIN phenotype measured by the ALF assay. Given that R-loops typically lead to double strand breaks and hyper-recombination due to interference with DNA replication forks [38], which has been observed in other DIS3 mutants [36,39], we also measured the frequency of Rad52-YFP foci in the DIS3 alleles. This analysis showed no increase in Rad52 foci, further suggesting that mutant-induced DNA damage is not a likely cause of genome instability (Figure 5(c)). Indeed, this is consistent with the lack of any sensitivity to genotoxic chemicals which damage DNA (Table 1; i.e. MMS, HU, camptothecin). Consequently, these data support our hypothesis emerging from the SGA, benomyl resistance, and dam1-1 suppression data, that mitotic spindle dysfunction could underlie the CIN phenotype of some DIS3 alleles. Previous studies have observed mitotic apparatus defects in ts-alleles of DIS3 [3], but did not link those defects to the exonuclease domain or to ongoing genome instability.
Perspective
Here we characterize a new allele of DIS3 in yeast, dis3E729K, which is orthologous to a mutation found in human cancers. Our results are consistent with this mutation, as well as dis3-ts, having mitotic defects, which are similar to those ascribed to the mtr17-1 allele of DIS3 previously shown to have abnormal mitotic spindles [3]. Our genome-wide screening data also supports this relationship with mitosis, and builds on these previous links of Dis3 to mitosis [3,4], by validating specific genetic dependencies of DIS3 mutants (e.g. with kinetochore function) in the context of a cancer-associated point mutation that exhibits genome instability. However, hundreds of gene products have altered levels of expression in DIS3 mutant alleles [24] and it remains difficult to confidently ascribe cellular phenotypes to specific gene expression changes. Regardless, our data suggests that exonuclease domain mutations seen in myelomas could potentially disrupt normal Dis3 functions to impact the mitotic apparatus and result in CIN.
More broadly, while various cellular pathways buffer genome stability directly (e.g. checkpoints and repair systems), genes with seemingly unrelated functions can influence DNA transactions such as repair, replication, or mitosis, if they have an impact on critical cellular processes (e.g. transcription, translation, and cellular energy status). Numerous RNA processing factors have now been implicated in genome maintenance and there are at least two possible models [40]. One model invokes the accumulation of unprocessed RNA leading to R-loops (DNA:RNA hybrids), which impair DNA replication causing DNA damage. The other model involves changes in gene expression that dysregulate more direct genome maintenance pathways such as DNA repair or mitotic function. In mutants of the DIS3 exonuclease domain, we do not find evidence of R-loops or DNA damage, suggesting that altered expression of genome maintenance factors is more likely driving genome instability. However, we cannot rule out a more direct role for Dis3 in mitosis. Regardless, the consequences of mitotic dysregulation by DIS3 mutants lead us to a speculative model in which specific myeloma-associated mutations impair normal cellular mitotic functions. In the longer term, through understanding the impact of cancer specific Dis3 mutations on molecular function, we expect that this information can be applied to a more complete description of DIS3 mutant function in myeloma.
Materials and methods
Yeast growth and strains
Yeast were grown in rich media at 30°C by standard procedures unless otherwise noted. The pDIS3-WT, dis3-EXO and dis3-ENDO alleles were created by plasmid shuffle [19]. Additional strains were taken from the collections described below (see Methods: ‘synthetic genetic array’). Strains used in this study are listed in Table S3.
Chromosome instability assays
For A-like faker (ALF), 9 independent colonies of each MATα mutant strain were inoculated overnight at permissive temperature in complete media. The next day, 100 μl of the saturated overnight cultures was mixed with 300 μL of a MATα mating tester strain, spun down, and resuspended in 100 μl distilled water. These MATα mixtures were then spotted on synthetic media lacking all supplements and only permitting prototrophic mated progeny to form colonies [31]. As well, the saturated overnight cultures were diluted by 1:100,000 and plated on complete media to ascertain colony viability. Plates were incubated at 30°C, and colonies counted once visible (typically after 3–4 days). Frequencies were calculated by dividing the number of colonies on depleted media by the colony count on complete media (which was multiplied by the dilution factor).
Synthetic genetic array
High-throughput screens were performed using the MATa non-essential gene deletion collection [21], a ts-allele collection [23], and the Decreased Abundance by mRNA Perturbation (DAmP) collection [22]. All array manipulation was done with a Singer RoToR HDA. Media and steps were modifed from Tong et al., 2004 [41]. Briefly, query strains were expanded on synthetic drop-out media selecting for the DIS3 allele at permissive temperature, then mated with the collection strains on rich media for one day at 30°C. Colonies were replicated to diploid-selective media plates for one day; this selection was repeated twice. Arrayed colonies were then replicated to sporulation media and allowed to grow at 25°C for 7 days. Sporulated arrays were expanded on haploid selection media in triplicate, and allowed to grow for roughly 30 hours; this haploid selection was then repeated a second time. Array plates were then replicated to haploid selective plates that carried additional selection for the collection alleles (‘single selection’) and incubated at 30°C for 3 days. Finally, each plate was concurrently replicated to single selection, as well as double selection (which also selects for the query allele; in this case, DIS3) and allowed to grow for one further day. Output arrays were scanned as plate images and colony size was analyzed using Balony Software [42]. Candidate hits were selected with a spot size difference (control vs experimental) cut-off of −0.2 and p-value < 0.05. Positive interactions were not analyzed for the purposes of this study.
Microscopy
Indicated strains were grown overnight at permissive temperature in appropriate drop-out selection media. The next day, the overnight cultures were diluted and imaged after 1.5 hours (once they had resumed growth), or shifted to the indicated temperatures for 2 further hours prior to imaging. Cultures were mounted on concanavalin-coated slides and imaged at 100x magnification on a fluorescent microscope (Leica) using Metamorph software (Molecular Devices). Final images were scored using Image J (rsbweb.nih.gov/ij/) as described [13,43]. For Rad52-YFP and Lsm1-GFP imaging, a minimum of 98 cells per replicate, >400 in total across all replicates were counted, imaged in DIC and YFP or GFP channels.
For chromosome spreads, log-phase cultures incubated at 30°C were spheroplasted for 45 minutes at 37°C, treated with 4% paraformaldehyde and 1% lipsol, and manually spread onto glass slides. The following day, slides were incubated for 30 minutes in blocking buffer (5% BSA, 0.2% milk in PBS) at room temperature in a humid chamber (which was also used for all subsequent incubation steps). Immunostaining was performed with S9.6 antibody (Kerafast) diluted in blocking buffer (1:1000) for 1 hour at room temperature. Slides were washed with PBS and incubated with a goat anti-mouse Cy3-conjugated secondary (Jackson ImmunoResearch 115–165-003) diluted 1:1000 in blocking buffer for 1 hour at 25°C. Slides were washed, mounted with 100 ng/ml DAPI in Vectashield mounting media (Vector Laboratories H-1000), and imaged as described above in DAPI and Cy3 channels. A minimum of 75 cells per replicate and >400 cells in total across all replicates was counted.
Northern blot
Yeast strains were grown to early-log phase in YPD at 30°C followed by a temperature shift to 37°C for two hours. Total RNA was extracted before and after the temperature shift as described previously by Hopper, Schultz, and Shapiro [44]. Briefly, cell pellets were re-suspended in equal volumes of ice-cold TSE buffer (0.01M Tris pH7.5, 0.01M EDTA, 0.1M sodium chloride) and TSE saturated phenol (pH = 7.4). The mixture was incubated for 20 minutes at 55°C and vortexed every 3 minutes. Phases were separated by centrifugation at 20,000g for 10 minutes and re-extracted with phenol. RNA was precipitated overnight in ethanol at −80°C.
Northern blotting was carried out as previously described [45]. 4 micrograms of total RNA was separated by electrophoresis on 10% TBE- 7M Urea gels and transferred onto a Hybond N+ membrane (Amersham). Gels were stained with 1ug/ml ethidium bromide to detect 5.8S and 5S rRNAs as loading controls. Membranes were cross-linked at 2400 J/m2 (UV Crosslinker, VWR). RNA was detected on the membrane using 10pmol/ml of a digoxigenin-labeled (DIG) probe. Probe17 sequence is 5′-GCGTTGTTCATCGATGC-3′ [46].
Fluorescence in situ hybridization
Experiments were performed as previously described [32]. Briefly, indicated strains were grown overnight at permissive temperature in appropriate drop-out selection media. The next day, the overnight cultures were diluted and, once in log phase, shifted to 37°C for 3 hours. Cells were fixed with 5% formaldehyde for 15 min. Hybridization was performed with a FITC–labeled oligo-dT, a Cy3-labelled probe for the 5.8S rRNA and Cy5-labelled probe for the ITS2 sequence [29 #1153, 46 #1297]; cultures were then washed and mounted with mounting media/DAPI for imaging.
Gene ontology and protein structure analysis
GO analysis was performed using the Generic GO term finder (http://go.princeton.edu/cgi-bin/GOTermFinder). Fold enrichments were calculated by computing the ratio of the number of genes associated with GO term in the hitlist and the number of genes associated with the GO term in the genome. The Dis3 crystal structure (PDB: 2VNU) [18] was visualized in DeepView (http://www.expasy.org/spdbv/) [47].
Funding Statement
This work was supported by the Canadian Institutes of Health Research [MOP 130231]; Canadian Institutes of Health Research [MOP 136982].
Acknowledgments
We acknowledge P. Hieter, D. Drubin, C. Schneider and C. Loewen for strains and protocols. KLM was supported by a Roman M. Babicki Fellowship in Medical Research and as a UBC Public Scholar. PCS was supported by a Canadian Institutes of Health Research grant (MOP 136982). AL was supported by a NSERC Canada Graduate Scholarship and BM by a Canadian Institutes of Health Research grant (MOP 130231). PCS is a Canadian Institutes of Health Research (CIHR) New Investigator and a Michael Smith Foundation for Health Research Scholar.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplemental data for this article can be accessed here.
References
- [1].Robinson SR, Oliver AW, Chevassut TJ, et al. The 3ʹ to 5ʹ exoribonuclease DIS3: from structure and mechanisms to biological functions and role in human disease. Biomolecules. 2015;5:1515–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Tomecki R, Drazkowska K, Kucinski I, et al. Multiple myeloma-associated hDIS3 mutations cause perturbations in cellular RNA metabolism and suggest hDIS3 PIN domain as a potential drug target. Nucleic Acids Res. 2014;42:1270–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Smith SB, Kiss DL, Turk E, et al. Pronounced and extensive microtubule defects in a Saccharomyces cerevisiae DIS3 mutant. Yeast. 2011;28:755–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Murakami H, Goto DB, Toda T, et al. Ribonuclease activity of Dis3 is required for mitotic progression and provides a possible link between heterochromatin and kinetochore function. PLoS One. 2007;2:e317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Snee MJ, Wilson WC, Zhu Y, et al. Collaborative control of cell cycle progression by the RNA exonuclease DIS3 and RAS is conserved across species. Genetics. 2016;203:749–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Ryu D, Kim HJ, Joung JG, et al. Comprehensive genomic profiling of IgM multiple myeloma identifies IRF4 as a prognostic marker. Oncotarget. 2016;7:47127–47133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Weissbach S, Langer C, Puppe B, et al. The molecular spectrum and clinical impact of DIS3 mutations in multiple myeloma. Br J Haematol. 2014. DOI: 10.1111/bjh.13256. [DOI] [PubMed] [Google Scholar]
- [8].Chapman MA, Lawrence MS, Keats JJ, et al. Initial genome sequencing and analysis of multiple myeloma. Nature. 2011;471:467–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Hanahan D, Weinberg RA.. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. [DOI] [PubMed] [Google Scholar]
- [10].Geigl JB, Obenauf AC, Schwarzbraun T, et al. Defining ‘chromosomal instability’. Trends Genet. 2008;24:64–69. [DOI] [PubMed] [Google Scholar]
- [11].Pfau SJ, Amon A. Chromosomal instability and aneuploidy in cancer: from yeast to man. EMBO Rep. 2012;13:515–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Stirling PC, Shen Y, Corbett R, et al. Genome destabilizing mutator alleles drive specific mutational trajectories in Saccharomyces cerevisiae. Genetics. 2014;196:403–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Stirling PC, Bloom MS, Solanki-Patil T, et al. The complete spectrum of yeast chromosome instability genes identifies candidate CIN cancer genes and functional roles for ASTRA complex components. PLoS Genet. 2011;7:e1002057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Stirling PC, Crisp MJ, Basrai MA, et al. Mutability and mutational spectrum of chromosome transmission fidelity genes. Chromosoma. 2012;121:263–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Ben-Aroya S, Coombes C, Kwok T, et al. Toward a comprehensive temperature-sensitive mutant repository of the essential genes of Saccharomyces cerevisiae. Mol Cell. 2008;30:248–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Bendl J, Stourac J, Salanda O, et al. PredictSNP: robust and accurate consensus classifier for prediction of disease-related mutations. PLoS Comput Biol. 2014;10:e1003440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Lionetti M, Barbieri M, Todoerti K, et al. A compendium of DIS3 mutations and associated transcriptional signatures in plasma cell dyscrasias. Oncotarget. 2015;6:26129–26141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Lorentzen E, Basquin J, Tomecki R, et al. Structure of the active subunit of the yeast exosome core, Rrp44: diverse modes of substrate recruitment in the RNase II nuclease family. Mol Cell. 2008;29:717–728. [DOI] [PubMed] [Google Scholar]
- [19].Schneider C, Leung E, Brown J, et al. The N-terminal PIN domain of the exosome subunit Rrp44 harbors endonuclease activity and tethers Rrp44 to the yeast core exosome. Nucleic Acids Res. 2009;37:1127–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Costanzo M, VanderSluis B, Koch EN, et al. A global genetic interaction network maps a wiring diagram of cellular function. Science. 2016;353 DOI: 10.1126/science.aaf1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Winzeler EA, Shoemaker DD, Astromoff A, et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. [DOI] [PubMed] [Google Scholar]
- [22].Breslow DK, Cameron DM, Collins SR, et al. A comprehensive strategy enabling high-resolution functional analysis of the yeast genome. Nat Methods. 2008;5:711–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Li Z, Vizeacoumar FJ, Bahr S, et al. Systematic exploration of essential yeast gene function with temperature-sensitive mutants. Nat Biotechnol. 2011;29:361–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Schneider C, Kudla G, Wlotzka W, et al. Transcriptome-wide analysis of exosome targets. Mol Cell. 2012;48:422–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Yoon JH, Choi EJ, Parker R. Dcp2 phosphorylation by Ste20 modulates stress granule assembly and mRNA decay in Saccharomyces cerevisiae. J Cell Biol. 2010;189:813–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Teixeira D, Sheth U, Valencia-Sanchez MA, et al. Processing bodies require RNA for assembly and contain nontranslating mRNAs. Rna. 2005;11:371–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Herrero AB, Moreno S. Lsm1 promotes genomic stability by controlling histone mRNA decay. Embo J. 2011;30:2008–2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Loll-Krippleber R, Brown GW. P-body proteins regulate transcriptional rewiring to promote DNA replication stress resistance. Nat Commun. 2017;8:558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Paul B, Montpetit B. Altered RNA processing and export lead to retention of mRNAs near transcription sites and nuclear pore complexes or within the nucleolus. Mol Biol Cell. 2016;27:2742–2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Vinton PJ, Weinert T. A slowed cell cycle stabilizes the budding yeast genome. Genetics. 2017;206:811–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Novoa CA, Ang JS, Stirling PC. The a-like faker assay for measuring yeast chromosome III stability. Methods Mol Biol. 2018;1672:1–9. [DOI] [PubMed] [Google Scholar]
- [32].Yuen KW, Warren CD, Chen O, et al. Systematic genome instability screens in yeast and their potential relevance to cancer. Proc Natl Acad Sci U S A. 2007;104:3925–3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Gomez-Gonzalez B, Garcia-Rubio M, Bermejo R, et al. Genome-wide function of THO/TREX in active genes prevents R-loop-dependent replication obstacles. Embo J. 2011. DOI: 10.1038/emboj.2011.206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Luna R, Jimeno S, Marin M, et al. Interdependence between transcription and mRNP processing and export, and its impact on genetic stability. Mol Cell. 2005;18:711–722. [DOI] [PubMed] [Google Scholar]
- [35].Gavalda S, Gallardo M, Luna R, et al. R-loop mediated transcription-associated recombination in trf4Delta mutants reveals new links between RNA surveillance and genome integrity. PLoS One. 2013;8:e65541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Chan YA, Aristizabal MJ, Lu PY, et al. Genome-wide profiling of yeast DNA:RNA hybrid prone sites with DRIP-chip. PLoS Genet. 2014;10:e1004288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Wahba L, Amon JD, Koshland D, et al. RNase H and multiple RNA biogenesis factors cooperate to prevent RNA:DNA hybrids from generating genome instability. Mol Cell. 2011;44:978–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Chang EY, Stirling PC. Replication fork protection factors controlling R-loop bypass and suppression. Genes (Basel). 2017;8 DOI: 10.3390/genes8010033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Tsanova B, Spatrick P, Jacobson A, et al. The RNA exosome affects iron response and sensitivity to oxidative stress. Rna. 2014;20:1057–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Chan YA, Hieter P, Stirling PC. Mechanisms of genome instability induced by RNA-processing defects. Trends Genet. 2014;30:245–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Tong AH, Lesage G, Bader GD, et al. Global mapping of the yeast genetic interaction network. Science. 2004;303:808–813. [DOI] [PubMed] [Google Scholar]
- [42].Young BP, Loewen CJ. Balony: a software package for analysis of data generated by synthetic genetic array experiments. BMC Bioinformatics. 2013;14:354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Hopper AK, Schultz LD, Shapiro RA. Processing of intervening sequences: a new yeast mutant which fails to excise intervening sequences from precursor tRNAs. Cell. 1980;19:741–751. [DOI] [PubMed] [Google Scholar]
- [45].Wu J, Bao A, Chatterjee K, et al. Genome-wide screen uncovers novel pathways for tRNA processing and nuclear-cytoplasmic dynamics. Genes Dev. 2015;29:2633–2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Allmang C, Mitchell P, Petfalski E, et al. Degradation of ribosomal RNA precursors by the exosome. Nucleic Acids Res. 2000;28:1684–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


