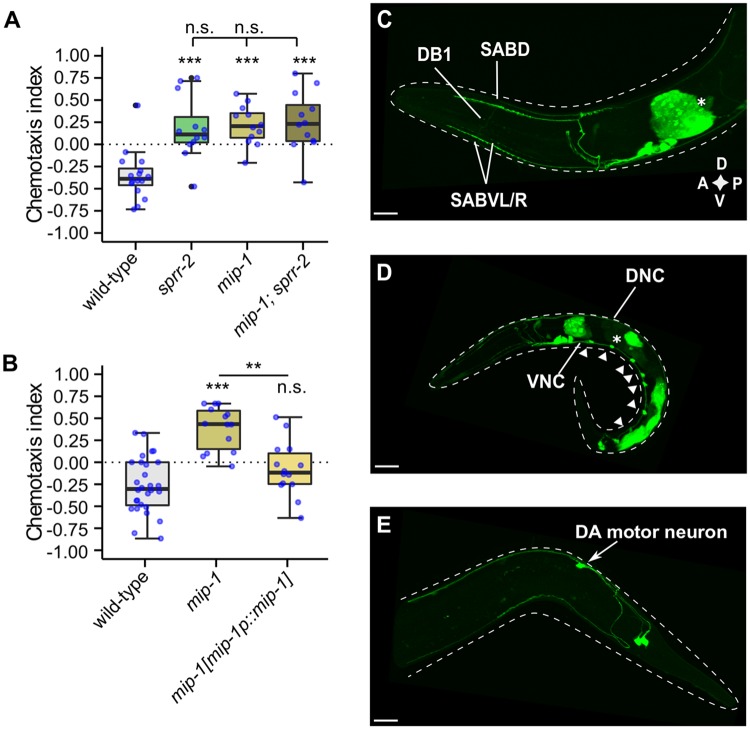
Fig 3. MIP-1 neuropeptides are required for gustatory plasticity.
(A-B) Gustatory plasticity of mip-1 mutant. Individual CIs, blue dots. Outliers, black dots. Boxplots indicate 25th (lower boundary), 50th (line), and 75th (upper boundary) percentiles. Whiskers show minimum and maximum values. Data were analyzed by one-way ANOVA with Tukey post-hoc test (n ≥ 8). *p<0.05; **p<0.005; ***p<0.001; n.s., not significant. (A) mip-1 mutants are defective in gustatory plasticity. A double mutant of mip-1 and sprr-2 has no additive learning defect. (B) The plasticity defect of mip-1 is rescued by restoring mip-1 expression under control of its promoter sequence [mip-1p::mip-1]. (C-E) Labeled confocal Z-stack projections showing expression of a reporter transgene for mip-1 [mip-1p::mip-1::sl2::gfp] in adult (C & E) or larval (D) hermaphrodites. Asterisks mark intestinal fluorescence resulting from the co-injection marker elt-2p::mCherry. Scale bars represent 15 μm. (C) mip-1 expression localizes to SABD, SABV and DB-1 motor neurons as well as around 4 additional unidentified neuron pairs in the head. (D) Expression was also found in dorsal and ventral nerve cords (DNC, VNC), and in motor neurons along the VNC (arrowheads). (E) mip-1 expression in the tail localizes to DA motor neurons, most likely DA8 or DA9, and an additional unidentified neuron pair. D, dorsal; V, ventral; A, anterior; P, posterior.