Abstract
This work extends the understanding of how toxic exposures to amphetamine (AMPH) adversely affect the immune system and lead to tissue damage. Importantly, it determines which effects of AMPH are and are not due to pronounced hyperthermia. Whole blood messenger RNA (mRNA) and whole blood and serum microRNA (miRNA) transcripts were identified in adult male Sprague-Dawley rats after exposure to toxic AMPH under normothermic conditions, AMPH when it produces pronounced hyperthermia, or environmentally-induced hyperthermia (EIH). mRNA transcripts with large increases in fold-change in treated relative to control rats and very low expression in the control group were a rich source of organ-specific transcripts in blood. When severe hyperthermia was produced by either EIH or AMPH, significant increases in circulating organ-specific transcripts for liver (Alb, Fbg, F2), pancreas (Spink1), bronchi/lungs (F3, Cyp4b1), bone marrow (Np4, RatNP-3b), and kidney (Cesl1, Slc22a8) were observed. Liver damage was suggested also by increased miR-122 levels in the serum. Increases in muscle/heart-enriched transcripts were produced by AMPH even in the absence of hyperthermia. Expression increases in immune-related transcripts, particularly Cd14 and Vcan, indicate that AMPH can activate the innate immune system in the absence of hyperthermia. Most transcripts specific for T-cells decreased 50–70% after AMPH exposure or EIH, with the noted exception of Ccr5 and Chst12. This is probably due to T-cells leaving the circulation and down-regulation of these genes. Transcript changes specific for B-cells or B-lymphoblasts in the AMPH and EIH groups ranged widely from decreasing ≈ 40% (Cd19, Cd180) to increasing 30 to 100% (Tk1, Ahsa1) to increasing ≥500% (Stip1, Ackr3). The marked increases in Ccr2, Ccr5, Pld1, and Ackr3 produced by either AMPH or EIH observed in vivo provide further insight into the initial immune system alterations that result from methamphetamine and AMPH abuse and could modify risk for HIV and other viral infections.
Introduction
Life-threatening hyperthermia is a serious toxic effect of amphetamines, including amphetamine (AMPH) and methamphetamine (METH), in both humans and laboratory animals [1–5]. In laboratory animals, the magnitude of hyperthermia correlates directly with the severity of neurotoxicity produced by AMPH and METH [6–9]. More recently, amphetamine-related damage to brain vasculature, liver, and kidney, and activation of the innate immune system have been implicated in exacerbating the neurotoxicity produced by amphetamines [10–14]. These effects are likely due, in part, to a hyperthermic response produced by amphetamines. This hyperthermic response is very similar to what occurs in heat stroke [15–18]. However, the effects of amphetamines and stress on neuroinflammation, and how this interaction may alter the neurotoxicity produced by amphetamines are complex and still being evaluated [19–23].
MicroRNAs (miRNAs) are small, 20–24 nucleotide-long endogenous non-coding RNAs that play an important role in the control of gene expression [24–26]. miRNA profiles vary by tissue and can be modulated by the onset and development of toxicities and diseases [27]. They are stable in biofluids, including blood, and the relative concentrations of specific circulating miRNAs are altered upon damage to different organs (e.g. liver, kidney, and heart) or after modulation of the immune system [27, 28]. Thus, several reports indicate the potential of circulating miRNAs as biomarkers of disease or toxicity [29, 30].
We hypothesize that many of the AMPH-produced adverse changes seen in the neuroimmune system [19, 20, 22, 23, 31, 32] will be reflected also in similar immune system changes in circulating blood, such as activation of the innate immune system. In addition, we thought it would be likely that both miRNA and mRNA transcripts would be present in the blood also as a result of AMPH-induced damage to liver, muscle, kidney, and pancreas. Detection of such miRNAs and mRNAs could provide additional biomarker candidates of the toxicity of AMPH and METH. Finally, it was hoped that the results of this study would help to clarify further how activation of immune system components in the circulating blood and damage of the brain vascular due to amphetamine may alter neuroinflammation, neurotoxicity, and infection [33–35].
The present study identifies the initial/early changes in mRNA and miRNA in circulating whole blood to address several issues, including the differences in the neurotoxicity and magnitude of the initial innate immune response between hyperthermic and non-hyperthermic exposures to AMPH, not determined in previous research. Messenger RNA and miRNA profiling was performed using both RNA sequencing (RNA-seq) and quantitative reverse transcription real-time polymerase chain reaction (RT-qPCR). An emphasis has been placed on determining the miRNA and mRNA expression changes, due to high level AMPH exposure, that affect immune function.
Portions of the mRNA RNA-seq data have been used in previous studies for three different research goals: 1) to develop RNA-seq outlier methodology [36]; 2) to evaluate the stability of RNA-seq gene expression [14]; and 3) to evaluate a multi-class classifier for RNA-seq data using a computational evolution system [37]. However, the miRNA expression data and their relevance to AMPH toxicity and the analysis of the mRNA expression changes have not been published before. As well, blood RNA-seq data related to hyperthermia alone (EIH) or a normothermic AMPH exposure have not been previously evaluated with respect to the entire transcriptome profiles and which transcripts are significantly changed by treatment. The present manuscript compares four treatment groups (new analysis) to better understand the range of amphetamine toxicities that can be produced. Because exposure to high doses of AMPH results in hyperthermia, inclusion of a normothermic AMPH group (kept in a 16°C environment to avoid hyperthermia) and an EIH group (which caused severe hyperthermia) enabled the determination of the non-hyperthermic- and hyperthermic-mediated effects of a neurotoxic AMPH exposure, which has not been previously reported. Finally, we have identified organ specific miRNA and mRNA transcripts in blood species that are potential biomarkers of toxicity (not previously reported).
Materials and methods
Animal dosing and sacrifice
This study was conducted in accordance with the Guide for the Care and Use of Laboratory Animals as adopted and promulgated by the National Institutes of Health. Animal procurement and testing was done under protocols E7295 and E7519 (issued to John Bowyer) approved by the NCTR institutional animal care and use committee (IACUC), accredited by NIH-OLAW. Nine- to ten-week-old (≈ 65 days) male Sprague-Dawley rats, ninety-five in total, were obtained from the Charles River Laboratories [Crl:CD(SD)], and received unique tail tattoos for identification after arrival at NCTR. Prior to testing, rats were housed two per cage with food and water available ad libitum, and kept on a daily 12 hr light cycle with lights on at 6:00 a.m. and off at 6:00 p.m. The environmental temperature (23°C) and humidity (53%) were controlled. The rats were tested between 12 and 13 weeks of age (≈ 87 days old).
Dosing occurred over a 6 h period starting at ≈ 8:00 a.m. and ending at ≈ 2:00 p.m. AMPH-exposed animals were given a sequential exposure to 5, 7.5, 10, and 10 mg AMPH/kg body weight (bw) subcutaneously with 2 hr between each dose. The AMPH hyperthermic (AMPH hyper) animals were dosed in a 23 to 24°C environment and AMPH normothermic (AMPH normo) animals were exposed in a 16°C environment. The D-amphetamine sulfate (Sigma-Aldrich, St. Louis, MO) was dissolved in normal saline for subcutaneous injection (1 ml/kg bw per injection). This AMPH dosing paradigm was very similar to an acute METH dosing paradigm that is often used to produce neurotoxicity [25, 26]. The saline control animals received 4 subcutaneous injections of 1 ml/kg bw each in a 23°C environment, and the EIH animals received 4 subcutaneous saline injections of 1 ml/kg bw each in a 39°C environment. In all groups, the behavior and body temperature of each rat was monitored hourly and until 3 hr after the last dose. The lethal effects of hyperthermia and hyperpyrexia can occur with AMPH when body temperatures exceed 41.5°C for more than 30 min. This was prevented by placing the animals unrestrained on crushed ice for 15 to 30 min in a clean plastic cage to drop their temperatures to below 40.0°C, as previously reported [38].
All the data for mRNA expression and miRNA expression determined by RT-qPCR for the four treatments were obtained from three replicates of the same experiment conducted over a 1.5-year period. The multiple replications were to ensure that any significant effects seen in transcript expression would be robust and reproducible over time. A forth additional experiment was conducted, which included only three dosing groups: saline given at 23°C (control), saline given in a cool 16°C environment, and AMPH hyper group. This final experiment was conducted approximately 4 months after the first three replicates. The cool control group was tested due to concerns that there might be a remote possibility of the cool environment affecting mRNA and miRNA profiles relative to the 23°C environment. Blood from the control and AMPH hyper groups were used in a previous publication to help determine the stability of RNAseq transcriptome (Group 4 data in Fig 1) and leukocyte subtype levels [36]. Total RNA from some of the control and AMPH hyper groups in this last experiment were used to generate a sufficient number of animals (n = 7 for both groups) to use RNAseq techniques to quantify blood miRNA levels. This was necessitated to due to depletion of the total RNA in the control and AMPH hyper groups in the first three replicate experiments, due to the RNAseq and RT-qPCR miRNA analysis processes.
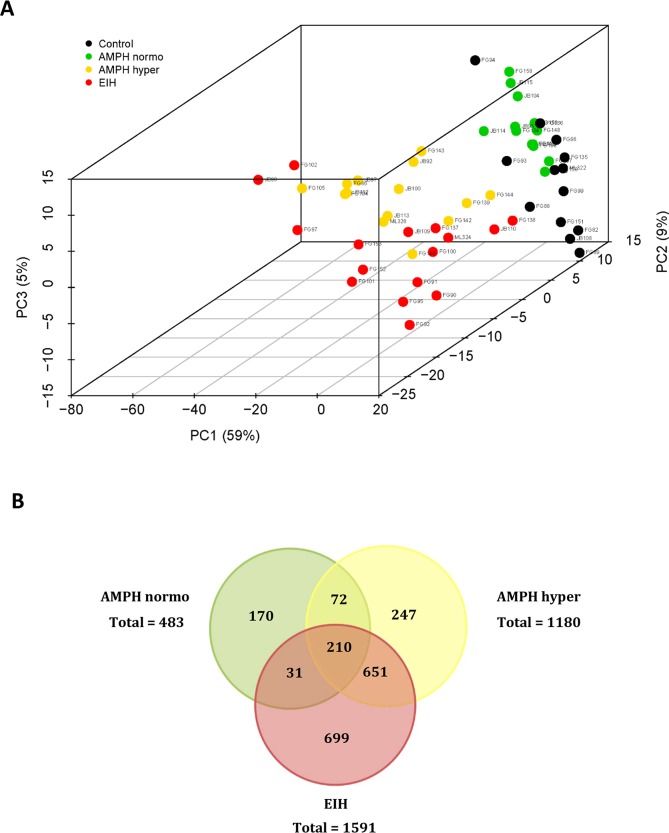
Fig 1. PCA and venn diagram analysis.
A. Principal component analysis score plot (3 principal components) of AMPH normo, AMPH hyper, EIH, and control treatment groups. B. Venn diagram showing the overlap of significant transcripts between the three main pairwise comparisons based on RNA-seq data analysis.
Collection and processing of cardiac blood
Rats were sacrificed 3 hr after the last dose, between 4:00 p.m. and 5:00 p.m., with a lethal dose of approximately 150 to 300 mg/kg i.p. injection of pentobarbital. This resulted in very slow and shallow respiration and no response to pain stimuli, within 3 min. At that point, 3 to 5 ml of cardiac blood was withdrawn from the heart using an 18-gauge needle attached to a 5 ml syringe, and the rats were then euthanatized by decapitation. Approximately 1 ml of the blood was immediately separated into three 300–400 μl aliquots, frozen on dry ice in cylindrical foil capsules, and stored at -70°C for later mRNA and miRNA profile analyses. One ml of the blood was placed on ice (1 to 2 hr) and serum was separated by centrifugation and stored at -70°C until analysis. For many of the controls and AMPH hyper animals, 1 ml of blood was placed in a 2 ml centrifuge tube with EDTA (10 mM final concentration) to determine blood cell type numbers, as previously described [14]. Data on the blood cell counts have been previously reported [14]. Any remaining blood was prepared as previously described for RNA isolation.
RNA isolation for mRNA and miRNA analyses
Total RNA isolation was performed using RNAzol BD (Molecular Research Center, Inc., www.mrcgene.com) [9] with modified procedures for frozen blood, as previously described [14]. Approximately 300 to 400 μl of frozen whole blood was used for RNA isolation. The final total cellular RNA recovered ranged from 5 to 25 μg and was stored at -70°C. Two to four μl of each aliquot were set aside for purity analysis using an Agilent 2200 TapeStation System (Agilent Technologies, Palo Alto, CA) and the quantity and purity of the RNA were assessed using a Nanodrop ND-8000 instrument (Thermo Scientific). RNA was isolated from 50 μl of serum using a MagMAx mirVana total RNA isolation kit and a King Fisher Flex purification system (Thermo Scientific).
A frozen aliquot of whole blood total RNA was shipped overnight on dry ice to Expression Analysis Inc. (EA; Durham, NC) for globin transcript removal and whole transcriptome sequencing. Alpha- and beta-globin mRNA were substantially depleted from total RNA samples using a GlobinClear-Mouse/Rat kit (Applied Biosystems, Foster City, CA) (for details see [14]). The final globin mRNA-depleted RNA samples were quantitated by spectrophotometry using a NanoDrop ND-8000 (Thermo Scientific, Waltham, MA).
mRNA quantitation
Transcript quantification in whole blood by RNA-seq profiling
Transcript expressions were determined by EA with their NGS sequencing methods, as previously described [14]. Briefly, EA used the globin mRNA-depleted RNA samples to create cDNA libraries using the TruSeq Stranded mRNA Sample Prep Kit (Illumina, San Diego, CA). Sequencing by synthesis methods, as implemented via Illumina technology, were used to generate the RNA-seq data. Fifty bp paired-end and strand-specific sequencing was performed on an Illumina platform generating 25–35 million reads per sample.
Quality control files contained read length and depth results (before and after clipping), presence of artifact/duplicate sequences, distribution of base quality, and base frequency by sample. Also, flow cell total yield, pass-filter reads, barcode quality, alignment summaries, and number of genes detected were determined. The basecall files were converted to fastq files using CASAVA 1.8.2. The fastq files were clipped using fastq-mcf with the parameters “—max-ns 4—qual-mean 25 -H -p 5 -q 7 -l 25” [39]. The fastq files were aligned to the rat Ensembl release 70 transcriptome (rn5) using bowtie 0.12.9 with the parameters “-e 500 –m 100 –chunkmbs 256”. The alignments were quantified using RSEM v 1.2.0 with no special parameters.
The RNA-seq data used in this study can be downloaded from NCBI GEO files GSE62368 and GSE64778. The IDs of the individual animals used in each group and assay are shown in Table 1. An additional set of samples was used to assess whether controls in a cool (16°C) environment responded similarly to controls in a 23°C environment. This set included three treatment groups: control (23°C environment), cool control (16°C environment), and AMPH hyper (S1 Table). There were no apparent differences in body temperature and expression profiles between the 16°C and 23°C control groups, including transcripts ascribed to organ damage or important immune-related genes (S1 and S2 Figs, and data not shown). Hence, all treatment comparisons, including of the AMPH normothermic group, were conducted relative to the 23°C control group.
Table 1. List of animal IDs included in mRNA and miRNA analyses.
| Animals used to determine RNA-seq profiles in whole blood | Animals used to determine the miRNA profiles in whole blood | |||
|---|---|---|---|---|
| Treatment Group | Individual Animal IDs a,b | Sample Size | Individual Animal IDs c | Sample Size |
|
Control
(saline given in a 23°C environment) |
ML322, JB108, FG82, FG88, FG89, FG93, FG94, FG98, FG99, FG135, FG136, FG150, FG151 | 13 (12) | ML322, FG82*, FG88*, FG89*, FG93*, FG98, FG135*, FG136*, FG150*, FG151* | 6 (10, 8*) |
| AMPH normo (AMPH given in a 16°C environment) | JB93, JB104, JB114, JB115, FG132, FG133, FG134, FG145, FG147, FG148, FG157, FG158, FG159 | 13 (13) | JB93, FG132*, FG134*, FG145*, FG147*, FG148*, FG157*, FG158*, FG159* | 6 (9, 8*) |
| AMPH hyper (AMPH given in a 23°C environment) | ML328, JB92, JB97, JB100, JB102, JB113, FG86, FG104, FG105, FG139, FG140, FG142, FG143, FG144 | 14 (12) | ML328, JB100, JB102*, FG86*, FG104*, FG105*, FG139*, FG140*, FG142*, FG143*, FG144* | 6 (11, 9*) |
|
EIH
(saline given in a 39°C environment) |
ML324, JB90, JB109, JB110, FG90, FG91, FG92, FG95, FG97, FG100, FG101, FG102, FG137, FG138, FG152, FG153 | 16 (15) | ML324, JB110, FG90*, FG91*, FG92*, FG95*, FG97, FG100*, FG101*, FG137*, FG138*, FG152*, FG153* | 6 (13, 10*) |
b Underlined IDs, blood samples were used in follow-up RT-qPCR analyses. The remaining samples had insufficient RNA to be analyzed by RT-qPCR.
c Bold IDs, blood samples used in TLDA and follow-up RT-qPCR analyses; IDs followed by *, serum samples used in RT-qPCR analyses. Serum samples were not available for all animals for which blood miRNAs were quantified by RT-qPCR, but the findings using all blood samples available or only the same sub-set of samples for which serum was also available were similar.
mRNA transcript quantification by quantitative reverse transcription–real-time polymerase chain reaction (RT-qPCR)
Reverse transcription of 500 ng of whole blood RNA was performed using random hexamers primers and SuperScript IV Reverse Transcriptase, followed by a digestion step at 37°C with E. coli RNase H (Invitrogen, Carlsbad, CA), following the manufacturer’s protocol. The cDNA was diluted 1:40 with nuclease-free water (Life Technologies, Foster City, CA) and used as a template in the qPCR assays. The expression of selected genes, identified as of interest based on the RNA-seq data, was quantified by qPCR in whole blood (n = 12-15/treatment group) using iQ SYBR Green Supermix and a CFX96 instrument (Bio-Rad Laboratories, Hercules, CA) and primers pairs designed using the NCBI Primer-BLAST software (S2 Table). The PCR cycling conditions were 1x 95°C for 10 min and 40x (95°C for 15 sec and 60°C for 1 min), followed by repetitive cycles of 5 sec starting at 65°C and incrementing in temperature by 0.5°C/cycle until 95°C to determine the melting curve data. Each primer pair yielded a single peak in the melting curve. Nuclease-free water (Life Technologies), instead of cDNA template, was added to the no-template control qPCR. Two independent qPCRs were performed to quantify the expression level of the endogenous control gene and the average of the two runs was used to normalize qPCR data.
miRNA quantitation
RT-qPCR methods for miRNA analysis
cDNA was synthesized by using a TaqMan MicroRNA Reverse Transcription Kit (Life Technologies). Two-hundred nanograms of whole blood RNA or 3μL serum RNA were reverse-transcribed by using MegaPlex RT primers 10x, Rodent Pool A (Applied Biosystems) and MultiScribe™ Reverse Transcriptase, following the manufacturer’s instructions. RT product was pre-amplified (12 cycles) with MegaPlex™ PreAmp primers 10x (Applied Biosystems), according to the manufacturer’s protocol.
The expression level of 381 miRNAs was quantified by qPCR in whole blood (n = 6/treatment group) using TaqMan Low Density Arrays (TLDA Rodent microRNA Array A, Applied Biosystems). The TLDA cards were run in an ABI 7900HT Fast Real-Time PCR System (Applied Biosystems), following the manufacturer’s recommended qPCR cycling parameters. The amplification curves were analyzed using Sequence Detection Systems (SDS) software, version 2.4.1 (Applied Biosystems). A subset of miRNAs (n = 9) was selected based on the TLDA screening data and quantified by RT-qPCR in whole blood (n = 9-13/treatment group) and in available serum samples from the same animals (n = 8-10/treatment group). cDNA was synthesized and pre-amplified as described above and used as a template in qPCR reactions, which further included TaqMan Universal PCR Master Mix II, No AmpErase UNG, and a miRNA-specific TaqMan Small RNA assay (Life Technologies). Data were collected using a 7900HT instrument (Applied Biosystems), following the manufacturer’s protocol. S3 Table lists the TaqMan Small RNA assays used.
RNAseq methods for miRNA analysis
A total of 14 RNA samples (control = 7 and AMPH hyper = 7) was sent to EA genomics (http://www.q2labsolutions.com/genomics-laboratoriesR). Their RIN number ranged from 7.0 to 9.3, with the control values of 8.3 ± 0.7 and AMPH hyper values of 8.4 ± 0.8 (mean ± SD). Sequencing libraries were created using an adaptation of the Illumina TruSeq Small RNA method. Briefly, single stranded adenylated DNA adapters were added to the 3’-hydroxyl group of the sample RNAs using T4 RNA Ligase 2, deletion mutant enzyme. This enzyme prevents ligation of the adapter to the 5’ end due to the absence of ATP. 5’ adapters were then added to the 5’-phosphate using T4 RNA Ligase in the presence of ATP. Following adapter ligation, single stranded cDNA was created by a reverse transcription reaction. The cDNA was then PCR amplified for 14 cycles using a common sequencing primer and an indexed primer that is unique to each sample. The cDNA libraries were then size selected using the Pippin HT to ensure sequencing of small RNAs from 15–35 base pairs. The resulting libraries were quantified, normalized, and pooled in preparation for sequencing. Ten million reads per sample were generated from the libraries. The methods developed were designed to greatly enrich the microRNA (miRNA) portion of an RNA fraction. This size-specific enrichment allows for an enhanced ability to detect small non-coding RNAs and possibly degraded fragments of longer RNA.
Data analysis
The RNA-seq by Expectation-Maximization (RSEM) [40] data generated by EA were rounded to produce count-based expression values. In total, 14,190 transcripts were included in the final analysis. Transcript-level outliers in each treatment group were identified by the iLOO [36]. and replaced with the trimmed mean. All reports of mean expression summarize DESeq2 normalized counts [41]. DESeq2 was also used to carry out differential expression analysis. Transcripts were deemed significant based on the following criteria: adjusted p-value (False Discovery Rate, FDR) < 0.05 and treatment mean must exceed DAFS global cutoff for low expression [14] or at least one of the two groups in pairwise comparison testing.
NormFinder [42] implemented in the R package NormqPCR [43] was used to identify the most stable miRNA from the TLDA screening data. NormFinder measures expression stability using model-based methods that account for both within and between-group variation. miR-148a was identified as the most stable miRNA and used as the endogenous control for miRNA data normalization. In previous work, Rpl22 was identified as the most stably expressed transcript in mRNA expression data [14] and hence used as the endogenous control for normalization of the mRNA qPCR data. The relative levels of miRNAs and mRNAs in whole blood quantified by RT-qPCR were calculated using the comparative ΔΔCt method [44]. One-way ANOVA was used to evaluate the significance of five planned pairwise comparisons: AMPH normo, AMPH hyper, and EIH to control, AMPH hyper to AMPH normo, and AMPH hyper to EIH. An FDR-adjusted p-value < 0.05 was considered statistically significant. All analyses were carried out in R (http://www.r-project.org).
Pathway analysis was performed using the Ingenuity Pathway Analysis (IPA, Ingenuity Systems, Redwood City, CA) software v7.1, which uses Fisher’s exact test to determine the likelihood of an association between significantly expressed transcripts and pathways of interest being due to random chance. Fold-change data (treated versus control) were uploaded to IPA to identify significantly enriched signaling and metabolic canonical pathways in the current data based on the Ingenuity Knowledge Base. Genes with an FDR-adjusted p-value < 0.05 and a │fold-change│ ≥ 2 versus control were analyzed. Pathways were evaluated based on a p-value to assess the significance of overlap between molecules from the dataset and literature and a z-score to assess whether a pathway is activated (z-score≥2.0) or inhibited (z-score≤-2.0). Ranked results from top canonical pathways meeting: p-value<0.01,│z-score│≥2.0, and containing at least three focus molecules, were queried for functional annotations and over-representation to facilitate biological interpretation of selected gene lists.
Results
Body temperatures and survival
Approximately 5% to 10% of the treated animals in both the AMPH hyper and EIH groups were not used in the study because they were moribund, had blood glucose levels less than 25 mg/dL, or did not become sufficiently hyperthermic. The remaining animals were considered suitable for mRNA and miRNA analysis of whole blood, and over 85% of these animals would have been expected to have a long-term survival after their exposures. There were no overwhelming differences between body temperature profiles of the AMPH normothermic and control groups (S1 Fig). Both the EIH and AMPH hyper groups had higher body temperatures compared to the AMPH normothermic and control groups, but there were no apparent differences in the temperature profiles between the EIH and AMPH hyper groups. However, the AMPH hyper group needed from two to five cooling “sessions” (depending on the individual rat) to protect against lethal hyperthermia, while the EIH group required only one or two sessions. Thus, the body temperature of the AMPH hyper animals dropped below 40°C two to five times between the 3rd and 9th hr after the start of dosing, while this only occurred once or twice in the EIH group.
RNA-seq differential expression analysis
Principal component analysis (PCA) was performed on ‘regularized log’ transformed RNA-seq data for the AMPH normo, AMPH hyper, EIH, control, and cool control treatment groups; the results are shown in S2 Fig. The cool control animals are surrounded by most of the main “core” of the control group. This suggests there is very little effect on the transcriptome due to a cool environment. It is also obvious that there are a few conspicuous outliers in several treatment groups. These outliers and the cool control group were removed and a PCA was subsequently performed on ‘regularized log’ transformed RNA-seq data for the AMPH normo, AMPH hyper, EIH, and control treatment groups.
A PCA score plot (3 principal components) is presented in Fig 1A. Treatment groups are summarized in S4 Table, where the mean and standard error of the DESeq2 normalized data are presented. A complete list of differentially expressed transcripts (i.e., FDR-adjusted p<0.05) for AMPH normo, AMPH hyper, and EIH relative to control is provided in S5 Table. The number of transcripts that were differentially expressed by at least 2-fold when comparing AMPH normo, AMPH hyper, and EIH to the control was: 483 (AMPH normo), 1180 (AMPH hyper), and 1591 (EIH) (Fig 1B). In addition, the list of differentially expressed transcripts between AMPH hyper relative to AMPH normo and EIH groups is provided in S6 Table. Of the differentially expressed transcripts, there were 820 and 182 differentially expressed transcripts with FC>2 when comparing AMPH hyper to AMPH normo and EIH, respectively.
Pathway analysis of the significant transcripts from the five pairwise comparisons was used to determine important changes produced by AMPH and/or hyperthermia (EIH). The canonical pathways significantly affected by treatment and the predicted direction of change for each pathway suggested a similar profile for the AMPH hyper and EIH groups (Fig 2). The number of pathways altered by the AMPH normo group relative to control was smaller than in the other two treatment groups. Moreover, their predicted activation/inhibition score tended to be smaller when compared to the controls (Fig 2 and S7 Table). Several pathways related to inflammation, including acute phase response and IL-6 and IL-8 signaling pathways, were activated in all treated groups relative to control. The LXR/RXR activation pathway was activated to a similar extent in AMPH hyper and EIH groups relative to control, while LPS/IL-1-mediated inhibition of RXR function pathway was inhibited in these two groups. Signaling pathways related to integrin-linked kinases and Rho GTPases, TFG-β, JAK/Stat, and actin cytoskeleton, all of which are involved in cell proliferation, migration, and apoptosis, were activated in the EIH group, and to a lesser extent in the AMPH hyper group, relative to control. In addition, the antioxidant action of vitamin C was inhibited, while the production of nitric oxide and reactive oxygen species in macrophages was activated (Fig 2 and S7 Table).
Fig 2. Heatmap displaying significant canonical pathways identified in the treatment groups of whole blood.
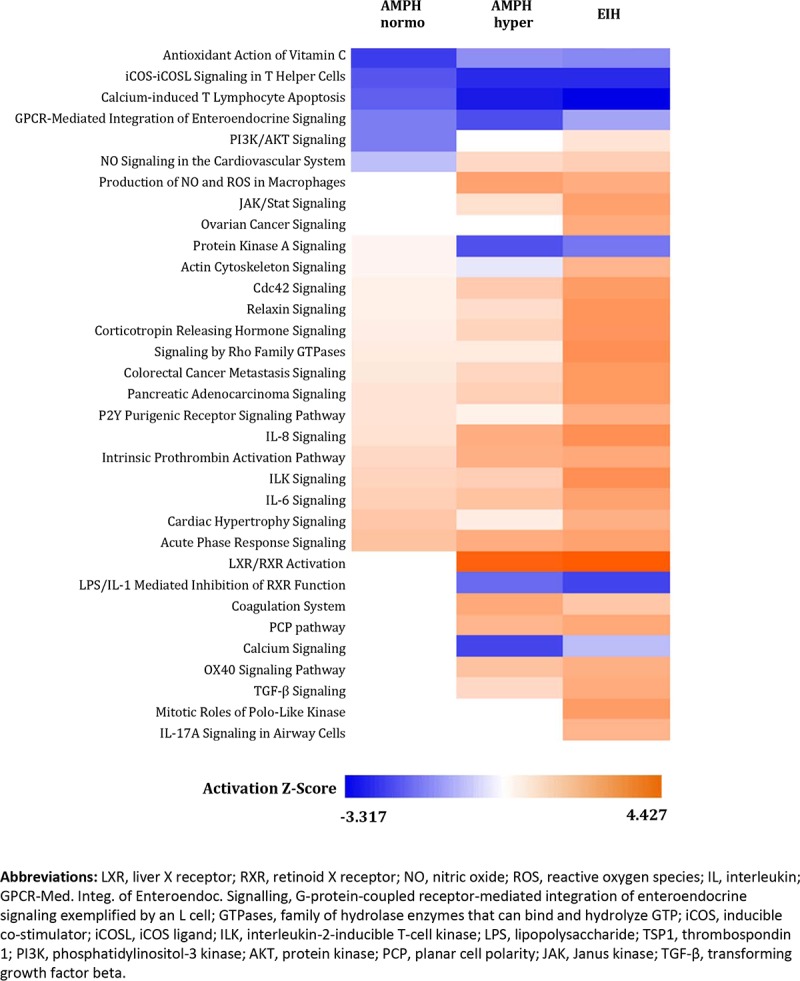
The treatment groups AMPH normo, AMPH hyper, and EIH groups relative to control, and of AMPH hyper relative to AMPH normo and EIH were analyzed using the Fisher’s exact test at p<0.01. The color gradient reflects the predicted direction of change for each pathway, based on regulation z-score, where blue represents inhibited and orange represents activated.
White blood cell-specific mRNAs among treatment groups
BioGPS [45] was used to determine the white blood cells (WBC) cell-type-specific genes shown in Fig 3 (monocytes), Fig 4 (T-cells), and Fig 5 (B-cells). Their origin was primarily determined using the available human and mouse transcriptomes. The transcriptome profiles in WBC cell-types for the mouse that are available in BioGPS appeared to correlate well with what was found in the corresponding human WBC cell-types. However, it is our opinion that the publicly available transcript expression references for blood or WBC cell-type in rats is incomplete at present, particularly compared to the human data. We hope these results will help build upon the current rat database for blood- and leukocyte-specific transcripts.
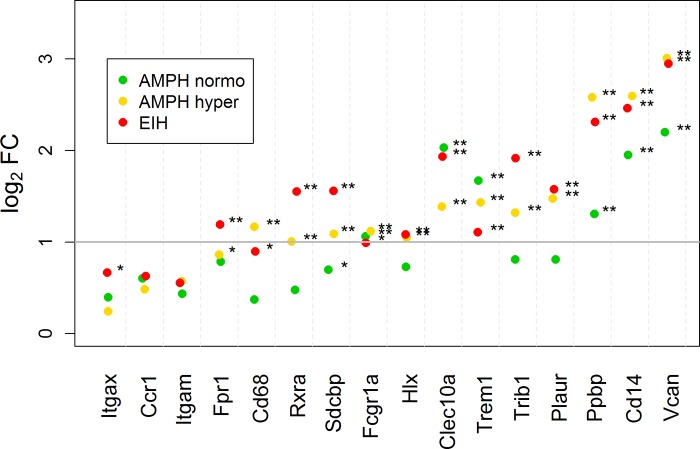
Fig 3. Scatterplot of fold-change expression for monocyte-specific mRNA transcripts in whole blood.
The log2 fold-changes for the AMPH normo, AMPH hyper, and EIH groups relative to control are shown. The horizontal gray bar indicates a 2-fold increase in expression compared to control. The subscripts * and ** indicate that the fold-change is significant and exceeds 1.5 and 2, respectively.
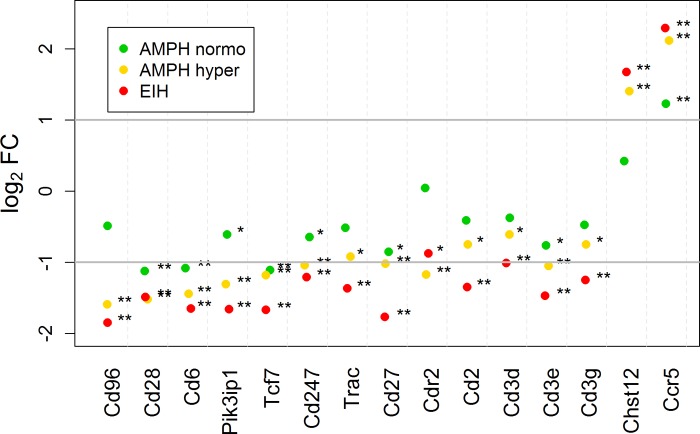
Fig 4. Scatterplot of fold-change expression for T-cell-specific mRNA transcripts in the whole blood.
The log2 fold-change for the AMPH normo, AMPH hyper, and EIH groups relative to control are shown. The horizontal gray bars indicate a 2-fold increase and decrease in expression compared to control. The subscripts * and ** indicate that the fold-change is significant and exceeds 1.5 and 2, respectively.
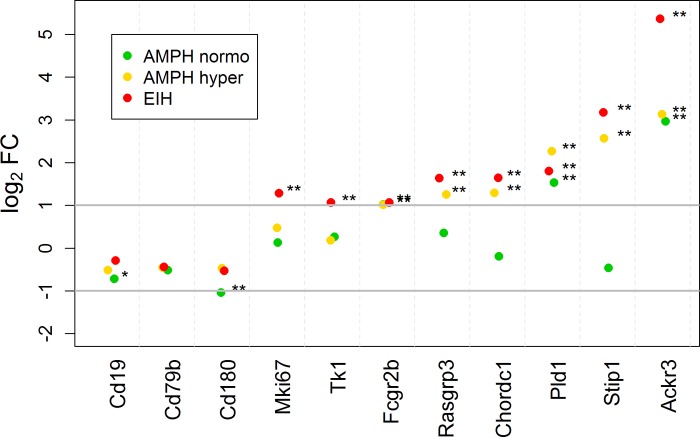
Fig 5. Scatterplot of fold-change expression for B-cell-specific mRNA transcripts in the whole blood.
The log2 fold-change for the AMPH normo, AMPH hyper, and EIH groups relative to control. The horizontal gray bars indicate a 2-fold increase and decrease in expression compared to control. The subscripts * and ** indicate that the fold-change is significant and exceeds 1.5 and 2, respectively.
Fig 3 presents the RNAseq log2 FC expression of monocyte-specific transcripts for AMPH normo, AMPH hyper, and EIH relative to control. For many transcripts, AMPH hyper and EIH were elevated by more than 1.5-fold. This is expected in the AMPH hyper group because monocyte counts in the blood were approximately double relative to control [14]. It is likely that this is also true for EIH; however, leukocyte sub-types counts were not determined for the EIH treatment group. In the EIH and AMPH hyper groups, some transcripts, such as Cd14, Clec10a, and Vcan, increased in expression by more than 4-fold. This greater fold-increase is most likely due to an up-regulation of these genes in monocytes already present in the blood, monocytes that entering the blood having a much higher expression of these genes, or a combination of these two possibilities. Our data suggest that hyperthermia was not required for AMPH to increase monocyte-specific transcripts, since several transcripts were significantly expressed by 2- to 4-fold in the AMPH normo group. It was not determined to what extent leukocyte sub-types numbers changed in this group. Interestingly, Clec10a and Trem1 were the only genes that increased to a greater extent in the AMPH normo group. Although not shown in Fig 3, the chemokine fractalkine receptor (Cx3cr1, found on monocytes, microglia, and maybe T-cells) paradoxically decreased in all three groups relative to control (S5 Table). RT-qPCR analysis confirmed the increases in fold-changes observed by RNA-seq for the monocyte membrane-associated transcript Vcan and Cd14 for the AMPH groups (S8 Table). RT-qPCR analysis also demonstrated increased expression in these groups relative to control for Ccr2, which is another monocyte membrane-associated transcript, but to which there was no alignment in the rat Ensembl release 70 transcriptome (rn5).
Fig 4 shows the magnitude of expression in T-cell-specific transcripts, relative to control, for AMPH normo, AMPH hyper, and EIH. Nearly all, except two transcripts that are not exclusively expressed in T-cells (Chst12 and Ccr5), were down-regulated by 2- to 4-fold in the AMPH hyper and EIH treatment groups. On the other hand, most transcripts specific to B-cells increased by 2- to 5-fold (Fig 5). Also, there was a more than 2-fold increase in immunoglobulin kappa constant (Igkc) and joining chain of multimeric IgA and IgM (Jchain) immunoglobulin transcripts in the AMPH hyper and EIH groups, but not in the AMPH normo group. These changes in both the T- and B-cells transcripts cannot be accounted for by changes in the blood cell count in the AMPH hyper group (and likely cannot for expression in the other two groups as well) (see Discussion). The reported fold-changes for Cd3d, Cd3g, Ccr5, and Ackr3, as determined by RT-qPCR, were similar to the fold-changes determined by RNA-seq (S8 Table).
Tissue-specific transcripts in whole blood
To detect organ or tissue damage, the RNA-seq data were filtered for transcripts with very low expression in the control group and much higher levels in AMPH normo, AMPH hyper, and EIH. This method also identified some of the heat-shock and stress proteins expected to increase with hyperthermia. The BioGPS web site [45] was used to determine the tissue or organ specificity for the selected transcripts. This was determined by examining the human and mouse transcriptomes. Tissue and organ specificity for rat tissue transcriptome was not heavily relied upon because the data for transcript origins in rats are not complete, particularly with respect to blood.
The top 245 transcripts with the greatest fold-change in the AMPH hyper group relative to the control group generated the initial list of candidate transcripts (S9 Table; transcripts listed in descending fold-change in EIH group versus control). Transcripts were retained if they had an NCBI gene symbol and one or more of the groups had a treatment mean > 13 (DAFS cutoff) (S5 Table). Of the top 35 transcripts, 23 were specific for liver (S9 Table). In fact, over half of the transcripts in the list were specific for liver. Genes present only in pancreas (Spink1), bronchi/lungs (Cyp4b1), bone marrow (Np4 [PRTN3 is analog humans] and RatNP-3b), and kidney (Cesl1 and Slc22a8) are also listed. Some of the genes present on the list are likely related to WBCs (e.g., Ackr3), B-cells, or miscellaneous tissues (Gpnmb, Ctsl1, and Gsg1). Transcripts enriched in heart/skeletal muscle should be interpreted with caution, due to the possibility of an artifact due to collection of terminal blood by cardiac puncture. RT-qPCR validated our RNA-seq findings for the non-liver-specific genes, including Ctsl1, F3, Gpnmb, and Gsg1 (S8 Table). However, the increased fold-changes in liver-specific transcripts (Alb, Crp, and Fgb) observed by RT-qPCR in the AMPH hyper group were smaller than those in the RNA-seq data, and not detected in the EIH group relative to control (S8 Table).
miRNA profiles in whole blood and serum
Of the 381 miRNAs screened in whole blood by RT-qPCR, 26 miRNAs and two small nuclear RNAs were found differentially expressed in one or more treatment groups relative to control (S10 Table). Twenty of the 26 miRNAs were increased in whole blood by more than 2-fold in the AMPH normo group relative to control, of which seven were also increased by a similar fold in the AMPH hyper group. In contrast, three miRNAs were decreased in whole blood of the EIH group relative to control (S10 Table). There was a relatively good agreement between the RT-qPCR method and the RNAseq method of quantifying miRNA levels in blood with the RNAseq method showing a tendency for lower fold-changes in general (Table 2 and S10 and S11 Tables). However, the comparison between the RNAseq small RNA and RT-qPCR methods can only be made with the control and AMPH hyper groups. Some miRNAs that appeared to be affected in the AMPH hyper group compared to control when evaluated by the RNAseq method had not been evaluated with the RT-qPCR methods (S11 Table).
Table 2. Fold-change of selected miRNAs in the whole blood and serum of AMPH normo, AMPH hyper, and EIH groups relative to control.
| AMPH normo | AMPH hyper | EIH | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Blood | Serum | Blood | Serum | Blood | Serum | ||||||
| miRNA | TLDA | qPCR | qPCR | TLDA | qPCR | RNAseq | qPCR | TLDA | qPCR | qPCR | |
| rno-miR-1-3p | 10.72* | 7.27* | 5.68 | 8.08* | 8.19* | 2.38 | 6.64 | 1.85 | 2.29 | 1.06 | |
| mmu-miR-122-5p | 2.34 | 2.51 | 26.60 | 8.78 | 5.44* | -1.28 | 22.22 | 1.57 | 1.77 | 35.52 | |
| mmu-miR-133a-3p | 15.26* | 9.76* | 4.62* | 10.78* | 9.60* | 5.50* | 4.44* | 2.50 | 2.85 | 1.30 | |
| mmu-miR-150-5p | 1.14 | -1.05 | 2.26 | -1.90 | -1.36 | -1.82 | 2.94* | -4.28* | -2.69* | 1.73 | |
| mmu-miR-204-5p | 4.43* | 3.50* | 1.54 | 2.45 | 2.43* | 1.07 | 3.15 | 1.22 | 1.31 | 2.74 | |
| mmu-miR-214-3p | 5.49* | 5.29* | 5.71* | 4.53* | 4.93* | 2.78 | 3.65* | 1.84 | 1.89 | 1.68 | |
| mmu-miR-223-3p | 3.64* | 2.41* | 1.86 | 1.06 | 1.48 | -1.15 | -1.01 | -1.88 | -1.28 | -1.55 | |
| mmu-miR-375-3p | 2.79* | 2.53* | 6.10* | 1.95 | 2.75* | 1.78* | 2.71 | 1.19 | 1.70 | 1.33 | |
| mmu-miR-429-3p | 25.80* | 2.86 | ND | 30.92* | 15.6* | 2.80* | ND | 16.08 | 3.43 | ND | |
a ND, not detected
Fold changes were assessed by TLDA screening (whole blood), RNAseq (control and AMPH hyper whole blood only), and RT-qPCR (whole blood and serum). Data are presented as mean fold-change relative to control.
*, p<0.05.
Consistent with the findings of increased skeletal muscle/heart-enriched mRNA transcripts in AMPH normo and AMPH hyper groups relative to control, the levels of several miRNAs reported to be up-regulated upon skeletal muscle damage or cardiac toxicity, including mmu-miR-1a-3p, rno-miR-1-3p, mmu-miR-133a-3p, and mmu-133b-3p, were increased by more than 4-fold in the whole blood of AMPH-treated animals (Table 2). Follow-up RT-qPCR of rno-miR-1-3p and mmu-miR-133a-3p confirmed their increased levels in whole blood, as well as in serum, although this effect was not significant (Table 2). In addition, mmu-miR-206-3p, which belongs to the same miRNA family, but was not represented in the RT-qPCR array plate, was found not to be expressed in any of the controls, but was detected in 6 of the 7 AMPH hyper animals by the RNAseq method (S11 Table.
Given the increased levels of liver-specific transcripts, such as Alb, Crp, and Fgb, in whole blood and the well-documented increases of circulating miR-122 upon liver injury, we quantified this miRNA in the whole blood and serum of study animals. Whole blood mmu-miR-122-5p was slightly increased by all treatments relative to control, with the effect being significant in the AMPH hyper group; however, a larger increase (> 22-fold) was observed in serum of all treated groups relative to control, although the large inter-animal variability resulted in non-statistical differences (Table 2). mmu-miR-429-3p was increased in blood by both AMPH exposures versus control; however, it was not detected in serum, suggesting that it is expressed in a blood cell rather than derived from a tissue (Table 2).
Discussion
The objectives of this study were attained in that 1) non-hyperthermic- and hyperthermic-mediated effects of high AMPH exposure on immune system and tissue damage were evaluated based on mRNA and miRNA expression; 2) organ-specific transcripts and miRNAs that relate to damage produced by AMPH or EIH/heat stroke were identified; and 3) the initial changes in mRNA and miRNA transcripts that could affect infection susceptibility were identified from AMPH exposure and EIH/heat stroke. The new data relevant to the first objective are important for translating animal experiments to AMPH toxicity in humans. We have previously reported the effects of AMPH on blood transcriptomics when life-threatening hyperthermia occurs [14]; however, the translational relevance of the changes that occur by AMPH when animals remain normothermic is important. It is not known what percentage of AMPH and METH abusers experience significant hyperthermic events during their lifetime, and it is not a common occurrence during the majority of stimulant exposures that an abuser has over his or her lifetime. However, if it only happens once in an abuser’s life-time, a hyperthermic crisis would likely result in longer lasting neurotoxicity and at least acute liver damage from what is known in studies with laboratory animals [8, 12, 38, 46].
For the AMPH hyper group, the changes in the expression of leukocyte-specific genes present in whole blood during the initial/early period (3 h after the toxic exposure regimen) are only partially explained by increases in monocytes (2-fold) and decreases in the total combined numbers of B-cells and T-cells (30%) in the circulating blood [14]. Similar significant changes in leukocyte-specific genes also occurred in the AMPH normo and EIH groups; however, we did not assess sub-types of leukocytes for these two groups. Overall, the observed transcript changes and alterations in leukocyte numbers are the initial indicators for activation of the innate immune system. We did not detect a significant increase in Il6 and Tnf in blood at the 3 hr time point, but they were also not observed to increase in meninges, choroid plexus, or brain tissues at this early time point in a previous study using the same toxic regimen [31]. However, at 24 h, both Il6 and Tnf had increased 2- to 3- fold in these tissues [31]. Thus, following either environmental heat exposure or exposure to AMPH, some classic signs of innate immune system activation, as represented by Il6 and Tnf, are present in laboratory animals.
We observed both an increased number of monocytes present in blood along with increased expression in cell surface proteins, such as Cd14, Ccr2, and Vcan, per monocyte present. The rapid rise in Ccl2 and possibly Ccl7 found in brain and other tissues produced by AMPH or METH [8, 19, 20, 22, 31] may be triggering this innate immune response through the Ccr2 receptors on monocytes and microglia [47–49]. In addition, damage-associated proteins from liver and other organs may be contributing to the innate immune response [50–53]. In conjunction with this, there is an apparent significant decrease in the number of circulating T-cells and possible down-regulation of some of their cell surface proteins per T-cell. This decrease may result from the rapid rise in Ccl17, a prime activator of T-cells [54, 55], transcripts in tissues such as brain, meninges and choroid plexus after neurotoxic exposures to either AMPH or METH [19, 20, 31]. Lastly, both an up-regulation and down-regulation of transcripts specific for B-cells was observed, which cannot be explained by changes in B-cell number in blood.
With respect to monocyte mRNA expression changes, the increases observed in Ccr2 and possibly Ccr5 would likely be expected to increase monocyte responsiveness to chemokines, while increases in Vcan and Cd14 would be expected to increase the ability of monocytes to detect bacterial and viral components that produce monocyte activation [47]. The data indicate that the expression for all four of these genes was increased by AMPH under normothermic conditions. In addition, activation of Trem1 amplifies monocyte-mediated inflammatory responses/activation triggered by bacterial and fungal infections by stimulating release of pro-inflammatory chemokines and cytokines. Its increase would be expected to result in monocyte activation and can result from AMPH exposure in the absence of hyperthermia. In contrast, unlike almost all the other monocyte transcripts, Cx3cr1 mRNA paradoxically decreased in all three groups relative to control. The decrease in the fractalkine receptor would be expected to decrease monocyte activation; however, this receptor is also on T-cells and natural killer (NK) cells and may just reflect the decrease in the number of T-cells circulating in blood along with a decrease in the transcription of T-cell-specific genes.
Even if all the decrease in total lymphocytes were due to loss of T-cells, this would only amount to a 1.7-fold decrease in T-cell-specific transcripts. Hence, the most plausible explanation for the increased B-cell transcripts is through increased numbers of transcripts per B-cell. There were seemingly anomalous increases in the expression levels of several genes within the T-cells. The unexpected increase in Ccr5 transcripts might be explained if it was up-regulated in the monocytes to overcome the lower levels of T-cells in blood. The unexpected increase in Chst12 transcript might have occurred due to entry of NK cells into the circulation or an upregulation of this gene in NK cells, since the cells have very high transcript levels of this gene in humans [45]. The large fold-increases in Pld1, Stip1, and Ackr3 in B-cells were significantly more than expected from any change in the number of B-cells in the blood.
The implications of a decrease in T-cells in the circulating blood, with a concomitant down-regulation of T-cell specific transcripts and an upregulation of B-cell transcript, are not clear. B-cells are known to be activated by T-cell-dependent and T-cell-independent mechanisms and it is possible that T-cell-independent B-cell activation is taking place following AMPH or hyperthermia. A fever-induced activation of B-cells could teleologically play a role in diseases in which high fever is produced, producing a polyclonal B-cell response. The upregulation of B-cell specific genes per B-cell would indicate an increase in B-cell activation, while the downregulation of T-cell-specific genes per T-cell would indicate a decrease. However, it is possible that many of the expression decreases in T-cell-specific genes are due to decreased numbers of T-cells within blood [14]. There were seemingly anomalous increases in the expression levels of several genes within the T- and B-cells. The more than doubling of immunoglobulin kappa constant (Igkc) and joining chain of multimeric IgA and IgM (Jchain) immunoglobulin transcripts in the AMPH hyper and EIH groups only indicates that extreme hyperthermia similar to heat stroke does activate B-cells.
In respect to T-cells, Saito et al. have previously reported the effects of METH on the immune status that have neurotoxic effects almost identical to AMPH in rodents [8, 38] in mice given a single 5 mg/kg dose or a ten day exposure to 5mg/kg per day [35]. Another mouse study by Martinez et al. used a similar single dose per day (but escalating by day with a 10 mg/kg ceiling) over a ten-day exposure, and observed signs of immunosuppression, reduced T-cell proliferation, and reduced resistance to fungal infection [56]. Some of the findings of the two mouse studies are similar to our results in rat. Saito et al. saw a decrease in the total number leukocytes of over 60% in mice, while our study saw leukocyte decreases ≤ 30% [14] and Martinez et al. observed signs of T-cell impairment. Our study results also indicated a significant down-regulation of genes important for T-cell immune function (e.g., Cd3d, Cd3e, and Cd3ge). Thus, all three studies saw signs of both very acute and more prolonged METH exposure adversely affecting T-cell function. There are significant differences between our study and the two mouse studies. Saito et al. saw a decrease in the NK cell activity acutely, while we found 4-fold-increase for the NK cell-specific transcript Chst12 when AMPH produced hyperthermia or with heat stroke, indicating increases in NK cells. Some of these differences are surely due to the previous studies examining expression in later stages of innate immune system activation, while our study observed changes in the initial stages resulting from AMPH exposure. In addition, some differences may be due to our AMPH exposure being significantly more acutely intense and toxic.
In vitro studies suggest that METH enhances HIV-1 replication in various HIV-1–permissive cells, including monocyte-derived macrophages and dendritic cells [57, 58]. METH has also been shown to increase replication of the feline immunodeficiency virus in astrocytes. Intriguingly, the effects of METH on HIV-1 replication in human CD4+ T-cells that are the primary targets of HIV-1 infection and replication in vivo remain largely unclear [57]. Our study found increases in several transcripts associated with HIV that have been reported to exacerbate the severity of HIV and other viral infections, occurred during AMPH exposure even under normothermic conditions. Since the number of macrophages increased by 2-fold, the 5-fold-increase in Ccr5, a co-receptor of HIV, in vivo indicates a slightly greater than 2-fold-increase in the expression of Ccr5. This same effect was previously observed in situ in human primary monocyte cultures, where it was mediated by both dopamine D1 and D2 receptor stimulation [59, 60]. These results are commensurate with our in vivo observation that Ccr5 expression can be upregulated by AMPH independent of hyperthermia. Also, our results indicate that hyperthermia alone, probably through activation of the innate immune system, can up-regulate Ccr5 expression to an even greater extent than just dopamine receptor stimulation. Ackr3, Pld1, and Ccr2 expression up-regulation had patterns very similar to Ccr5 and further indicate that AMPH and EIH can exacerbate HIV-1 infection [61–63].
Many transcripts enriched in liver (e.g. Alb, Crp, and Fgb), which had little expression in the blood of controls and AMPH normo rats, were highly up-regulated in the AMPH hyper and EIH groups. This is due to hyperthermia damaging the liver [12, 13, 64], and almost certainly results from degenerating hepatocytes releasing mRNA into the circulating blood. The increase of serum miR-122 in these animals further suggests liver damage. Detection of up-regulated transcripts specific for pancreas (Spink1), bronchi/lungs (Cyp4b1 and F3), and kidney (Cesl1 and Slc22a8) indicate that damaged or degenerating cells from these organs are also releasing mRNA, but to a lesser extent. Such increases in mRNA specific for organ tissues was not detected in the AMPH normo group, which shows that this effect is related to hyperthermia produced by AMPH. It should be noted that the fold-changes observed in RT-qPCR data, especially with regards to liver-specific transcripts, were much lower than those observed in RNA-seq data for EIH and, to a lesser extent, AMPH hyper. This suggests that heat may cause modifications to the RNA molecules and interfere with the ability to detect them.
Some transcripts present in the whole blood of AMPH hyper and EIH rats, which were not contained within WBCs or RBCs, appear to be more likely to breakdown or undergo chemical reactions and/ or degradation so that they cannot be quantified using RT-qPCR. We postulate this because RNA-seq techniques can detect tremendous increases in the levels of several liver-specific transcripts (Alb, Crp, and Fgb), while these transcript elevations are less pronounced or not detected at all with RT-qPCR methods. This inability to quantify mRNA levels with RT-qPCR in the two hyperthermic group may be because their body temperature remained above 40°C for extended time periods, which will greatly increase tissue damage and reactive oxygen species (ROS) [8]. This could in turn increase the ability of the mRNA to react with constituents in the plasma and somehow interfere with their detection by RT-qPCR. This effect was greater in the EIH group than the AMPH hyper group, possibly because the EIH group had even longer continuous periods where the body temperature was above 40°C. At the miRNA level, the most changes detected by RT-qPCR were in the AMPH normo group, followed by the AMPH hyper group; however, it is perplexing that the RT-qPCR methods of quantification show the same or higher fold-changes when compared to the RNAseq method in several of the miRNAs affected as a result of the AMPH hyper exposure. Nonetheless, these results indicate that physiological factors such as hyperthermia may alter the ability to detect mRNA and miRNA in blood.
Several miRNAs were found modulated in the blood of AMPH-exposed animals versus the controls. The levels of miR-1, miR-133a, and miR-206, which play a role in skeletal muscle development, function, and disease [65], were increased by AMPH regardless of the thermic conditions. In addition, several miRNAs reported to affect aspects of immune and vascular function were also found increased, including miR-429, miR-100, and miR-214, [66–68]. Many of these miRNA changes were observed in both AMPH normo and AMPH hyper groups. Such changes may be related to regulation of leukocyte function.
Conclusions
In this study the non-hyperthermic- and hyperthermic-mediated effects of a high dose AMPH were determined in the transcriptome of circulating blood. AMPH alone, in the absence of hyperthermia, was found capable of significantly affecting the immune status and leukocyte subtype-specific transcriptomics. The AMPH-dependent changes in leukocyte cell-type, along with the mRNA and miRNA changes that we observed, clearly manifest as early indicators of innate immune system activation in response to severe hyperthermia. Interestingly, extreme hyperthermia, like that producing heat-stroke, had very similar effects. The changes in leukocyte-specific transcripts also indicated that either AMPH (even when it does not produce hyperthermia) or extreme hyperthermia could increase the susceptibility to viral infection and/or accelerate the debilitating effects of viral infection. In addition, organ-specific transcripts that relate to damage produced by AMPH when it produces hyperthermia or EIH/heat stroke are present in blood acutely following exposure. This indicates that overt tissue damage that occurs during exposure to amphetamines is amplified greatly by hyperthermia.
Supporting information
This figure shows that rat body temperature is minimally affected by amphetamine (AMPH) when the room temperature is 16°C, but that severe hyperthermia usually occurs when AMPH is given and the room temperature is 23.5°C. It also shows that rats do not become hypothermic when given saline when the room temperature is 16°C.
(PDF)
This PCA plot shows that the Cool Saline (given saline in a 16°C rather than 23.5°C normal room temperature) animal transcriptome in blood was virtually identical to the normal room temperature saline animals. The red symbols for the Cool Saline Controls are embedded and obscured among the black symbols representing the Saline Controls (room temperature). It also shows the outlier in the Saline Controls and other groups that were removed from analysis and are not present in Fig 1A shown in the manuscript.
(PDF)
(DOCX)
(DOCX)
(DOCX)
(XLSX)
1st Sheet: Genes differentially expressed between AMPH normo and Control at FDR-adj-p < 0.05; 2nd Sheet: Genes differentially expressed between AMPH hyper and Control at FDR-adj-p < 0.05; 3rd Sheet: Genes differentially expressed between EIH and Control at FDR-adj-p < 0.05.
(XLSX)
1st Sheet: Genes differentially expressed between AMPH hyper and AMPH normo at FDR-adj-p < 0.05; 2nd Sheet: Genes differentially expressed between AMPH hyper and EIH at FDR-adj-p < 0.05.
(XLSX)
Only canonical pathways that had a p-value<0.01, │z-score│≥2. 0, and that contained at least three focus molecules for at least one of the treatment groups relative to control are listed.
(DOCX)
Data are presented as mean fold-change relative to control. *, p<0.05.
(DOCX)
(XLSX)
Data are presented as mean fold-change relative to control. * p<0.05.
(DOCX)
(DOCX)
Data Availability
All relevant data are within the paper and its Supporting Information files. The RNA-seq data are available from NCBI GEO database (GSE62368, GSE64778, GSE115835).
Funding Statement
Research animals, their housing, and reagents were funded under NCTR/FDA protocols E7295 and E7519.
References
- 1.Davis WM, Bedford JA, Buelke JL, Guinn MM, Hatoum HT, Waters IW, et al. Acute toxicity and gross behavioral effects of amphetamine, four methoxyamphetamines, and mescaline in rodents, dogs, and monkeys. Toxicol Appl Pharmacol. 1978;45(1):49–62. . [DOI] [PubMed] [Google Scholar]
- 2.Davis WM, Hatoum HT, Waters IW. Interactions of catecholaminergic receptor blockers with lethal doses of amphetamine or substituted amphetamines in mice. Res Commun Chem Pathol Pharmacol. 1978;21(1):27–36. . [PubMed] [Google Scholar]
- 3.Kalant H, Kalant OJ. Death in amphetamine users: causes and rates. Can Med Assoc J. 1975;112(3):299–304. [PMC free article] [PubMed] [Google Scholar]
- 4.Zalis EG, Lundberg GD, Knutson RA. The pathophysiology of acute amphetamine poisoning with pathologic correlation. J Pharmacol Exp Ther. 1967;158(1):115–27. . [PubMed] [Google Scholar]
- 5.Zalis EG, Parmley LF Jr. Fatal Amphetamine Poisoning. Arch Intern Med. 1963;112(6):822–6. 10.1001/archinte.1963.03860060060004 [DOI] [PubMed] [Google Scholar]
- 6.Bowyer JF, Davies DL, Schmued L, Broening HW, Newport GD, Slikker W Jr., et al. Further studies of the role of hyperthermia in methamphetamine neurotoxicity. J Pharmacol Exp Ther. 1994;268(3):1571–80. . [PubMed] [Google Scholar]
- 7.Bowyer JF, Tank AW, Newport GD, Slikker W Jr., Ali SF, Holson RR. The influence of environmental temperature on the transient effects of methamphetamine on dopamine levels and dopamine release in rat striatum. J Pharmacol Exp Ther. 1992;260(2):817–24. . [PubMed] [Google Scholar]
- 8.Bowyer JFH, J.P. Amphetamine- and methamphetamine-induced hyperthermia: Implications of the effects produced in brain vasculature and peripheral organs to forebrain neurotoxicity. Temperature. 2014;1(3):172–82. 10.4161/23328940.2014.982049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O'Callaghan JP, Miller DB. Neurotoxicity profiles of substituted amphetamines in the C57BL/6J mouse. J Pharmacol Exp Ther. 1994;270(2):741–51. . [PubMed] [Google Scholar]
- 10.Bowyer JF, Ali S. High doses of methamphetamine that cause disruption of the blood-brain barrier in limbic regions produce extensive neuronal degeneration in mouse hippocampus. Synapse. 2006;60(7):521–32. 10.1002/syn.20324 . [DOI] [PubMed] [Google Scholar]
- 11.Bowyer JF, Sarkar S, Tranter KM, Hanig JP, Miller DB, O'Callaghan JP. Vascular-directed responses of microglia produced by methamphetamine exposure: indirect evidence that microglia are involved in vascular repair? J Neuroinflammation. 2016;13(1):64 10.1186/s12974-016-0526-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Halpin LE, Yamamoto BK. Peripheral ammonia as a mediator of methamphetamine neurotoxicity. J Neurosci. 2012;32(38):13155–63. 10.1523/JNEUROSCI.2530-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Levi MS, Patton RE, Hanig JP, Tranter KM, George NI, James LP, et al. Serum myoglobin, but not lipopolysaccharides, is predictive of AMPH-induced striatal neurotoxicity. Neurotoxicology. 2013;37C:40–50. Epub 2013/04/24. S0161-813X(13)00055-7 [pii] 10.1016/j.neuro.2013.04.003 . [DOI] [PubMed] [Google Scholar]
- 14.Bowyer JF, Tranter KM, Hanig JP, Crabtree NM, Schleimer RP, George NI. Evaluating the Stability of RNA-Seq Transcriptome Profiles and Drug-Induced Immune-Related Expression Changes in Whole Blood. PloS one. 2015;10(7):e0133315 10.1371/journal.pone.0133315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gomez CR. Disorders of body temperature. Handb Clin Neurol. 2014;120:947–57. 10.1016/B978-0-7020-4087-0.00062-0 . [DOI] [PubMed] [Google Scholar]
- 16.Huang JJ, Desai C, Singh N, Sharda N, Fernandes A, Riaz IB, et al. Summer syncope syndrome redux. Am J Med. 2015;128(10):1140–3. 10.1016/j.amjmed.2015.05.016 . [DOI] [PubMed] [Google Scholar]
- 17.Leon LR, Bouchama A. Heat stroke. Compr Physiol. 2015;5(2):611–47. 10.1002/cphy.c140017 . [DOI] [PubMed] [Google Scholar]
- 18.Heled Y, Fleischmann C, Epstein Y. Cytokines and their role in hyperthermia and heat stroke. J Basic Clin Physiol Pharmacol. 2013;24(2):85–96. 10.1515/jbcpp-2012-0040 . [DOI] [PubMed] [Google Scholar]
- 19.O'Callaghan JP, Kelly KA, VanGilder RL, Sofroniew MV, Miller DB. Early activation of STAT3 regulates reactive astrogliosis induced by diverse forms of neurotoxicity. PloS one. 2014;9(7):e102003 10.1371/journal.pone.0102003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O'Callaghan JP, Sriram K, Miller DB. Defining "neuroinflammation". Ann N Y Acad Sci. 2008;1139:318–30. 10.1196/annals.1432.032 . [DOI] [PubMed] [Google Scholar]
- 21.Sriram K, Miller DB, O'Callaghan JP. Minocycline attenuates microglial activation but fails to mitigate striatal dopaminergic neurotoxicity: role of tumor necrosis factor-alpha. Journal of neurochemistry. 2006;96(3):706–18. Epub 2006/01/13. JNC3566 [pii] 10.1111/j.1471-4159.2005.03566.x . [DOI] [PubMed] [Google Scholar]
- 22.Thomas DM, Francescutti-Verbeem DM, Liu X, Kuhn DM. Identification of differentially regulated transcripts in mouse striatum following methamphetamine treatment—an oligonucleotide microarray approach. Journal of neurochemistry. 2004;88(2):380–93. . [DOI] [PubMed] [Google Scholar]
- 23.Thomas DM, Walker PD, Benjamins JA, Geddes TJ, Kuhn DM. Methamphetamine neurotoxicity in dopamine nerve endings of the striatum is associated with microglial activation. J Pharmacol Exp Ther. 2004;311(1):1–7. 10.1124/jpet.104.070961 . [DOI] [PubMed] [Google Scholar]
- 24.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–97. Epub 2004/01/28. . [DOI] [PubMed] [Google Scholar]
- 25.Shivdasani RA. MicroRNAs: regulators of gene expression and cell differentiation. Blood. 2006;108(12):3646–53. Epub 2006/08/03. 10.1182/blood-2006-01-030015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10(2):126–39. Epub 2009/01/24. 10.1038/nrm2632 . [DOI] [PubMed] [Google Scholar]
- 27.Siddeek B, Inoubli L, Lakhdari N, Rachel PB, Fussell KC, Schneider S, et al. MicroRNAs as potential biomarkers in diseases and toxicology. Mutat Res Genet Toxicol Environ Mutagen. 2014;764–765:46–57. Epub 2014/02/04. 10.1016/j.mrgentox.2014.01.010 . [DOI] [PubMed] [Google Scholar]
- 28.Yu HW, Cho WC. The role of microRNAs in toxicology. Arch Toxicol. 2015;89(3):319–25. Epub 2015/01/15. 10.1007/s00204-014-1440-2 . [DOI] [PubMed] [Google Scholar]
- 29.Correia CN, Nalpas NC, McLoughlin KE, Browne JA, Gordon SV, MacHugh DE, et al. Circulating microRNAs as Potential Biomarkers of Infectious Disease. Front Immunol. 2017;8:118 10.3389/fimmu.2017.00118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laterza OF, Lim L, Garrett-Engele PW, Vlasakova K, Muniappa N, Tanaka WK, et al. Plasma MicroRNAs as sensitive and specific biomarkers of tissue injury. Clin Chem. 2009;55(11):1977–83. 10.1373/clinchem.2009.131797 . [DOI] [PubMed] [Google Scholar]
- 31.Bowyer JF, Patterson TA, Saini UT, Hanig JP, Thomas M, Camacho L, et al. Comparison of the global gene expression of choroid plexus and meninges and associated vasculature under control conditions and after pronounced hyperthermia or amphetamine toxicity. BMC Genomics. 2013;14:147 Epub 2013/03/19. 1471-2164-14-147 [pii] 10.1186/1471-2164-14-147 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hess A, Desiderio C, McAuliffe WG. Acute neuropathological changes in the caudate nucleus caused by MPTP and methamphetamine: immunohistochemical studies. J Neurocytol. 1990;19(3):338–42. Epub 1990/06/01. . [DOI] [PubMed] [Google Scholar]
- 33.Chang L, Ernst T, Speck O, Grob CS. Additive effects of HIV and chronic methamphetamine use on brain metabolite abnormalities. Am J Psychiatry. 2005;162(2):361–9. 10.1176/appi.ajp.162.2.361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Soontornniyomkij V, Umlauf A, Soontornniyomkij B, Batki IB, Moore DJ, Masliah E, et al. Lifetime methamphetamine dependence is associated with cerebral microgliosis in HIV-1-infected adults. J Neurovirol. 2016;22(5):650–60. 10.1007/s13365-016-0441-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saito M, Terada M, Kawata T, Ito H, Shigematsu N, Kromkhun P, et al. Effects of single or repeated administrations of methamphetamine on immune response in mice. Exp Anim. 2008;57(1):35–43. . [DOI] [PubMed] [Google Scholar]
- 36.George NI, Bowyer JF, Crabtree NM, Chang CW. An Iterative Leave-One-Out Approach to Outlier Detection in RNA-Seq Data. PloS one. 2015;10(6):e0125224 10.1371/journal.pone.0125224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Crabtree NM, Moore JH, Bowyer JF, George NI. Multi-class computational evolution: development, benchmark evaluation and application to RNA-Seq biomarker discovery. BioData Min. 2017;10:13 10.1186/s13040-017-0134-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bowyer JF, Thomas MT, Schmued LC, Ali SF. Brain region-specific neurodegenerative profiles showing the relative importance of amphetamine dose, hyperthermia, seizures and the blood-brain barrier. Ann NY Acad Sci. 2008;1139:127–39. 10.1196/annals.1432.005 [DOI] [PubMed] [Google Scholar]
- 39.Aronesty E. Comparison of sequencing utility programs. Open Bioinform J. 2013;7:1–8. [Google Scholar]
- 40.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323 10.1186/1471-2105-12-323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Love M, Huber H, Anders S. Moderated estimation of fold change and dispersion for RNA-Seq data with DESeq2. bioRxiv. Preprint Published online February. 2014;19:2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Andersen CL, Jensen JL, Orntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64(15):5245–50. 10.1158/0008-5472.CAN-04-0496 . [DOI] [PubMed] [Google Scholar]
- 43.Perkins JR, Dawes JM, McMahon SB, Bennett DL, Orengo C, Kohl M. ReadqPCR and NormqPCR: R packages for the reading, quality checking and normalisation of RT-qPCR quantification cycle (Cq) data. BMC Genomics. 2012;13:296 10.1186/1471-2164-13-296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–8. 10.1006/meth.2001.1262 . [DOI] [PubMed] [Google Scholar]
- 45.Wu C, Orozco C, Boyer J, Leglise M, Goodale J, Batalov S, et al. BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol. 2009;10(11):R130 10.1186/gb-2009-10-11-r130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Levi MS, Divine B, Hanig JP, Doerge DR, Vanlandingham MM, George NI, et al. A comparison of methylphenidate-, amphetamine-, and methamphetamine-induced hyperthermia and neurotoxicity in male Sprague-Dawley rats during the waking (lights off) cycle. Neurotoxicol Teratol. 2012;34(2):253–62. Epub 2012/02/01. S0892-0362(12)00020-7 [pii] 10.1016/j.ntt.2012.01.007 . [DOI] [PubMed] [Google Scholar]
- 47.Akira S, Misawa T, Satoh T, Saitoh T. Macrophages control innate inflammation. Diabetes, obesity & metabolism. 2013;15 Suppl 3:10–8. 10.1111/dom.12151 . [DOI] [PubMed] [Google Scholar]
- 48.Di Santo JP. Staying innate: transcription factor maintenance of innate lymphoid cell identity. Immunological reviews. 2014;261(1):169–76. 10.1111/imr.12202 . [DOI] [PubMed] [Google Scholar]
- 49.Zhao S, Li F, Leak RK, Chen J, Hu X. Regulation of Neuroinflammation through Programed Death-1/Programed Death Ligand Signaling in Neurological Disorders. Front Cell Neurosci. 2014;8:271 10.3389/fncel.2014.00271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lacour S, Gautier JC, Pallardy M, Roberts R. Cytokines as potential biomarkers of liver toxicity. Cancer Biomark. 2005;1(1):29–39. . [DOI] [PubMed] [Google Scholar]
- 51.Hirsiger S, Simmen HP, Werner CM, Wanner GA, Rittirsch D. Danger signals activating the immune response after trauma. Mediators Inflamm. 2012;2012:315941 10.1155/2012/315941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kang R, Lotze MT, Zeh HJ, Billiar TR, Tang D. Cell Death and DAMPs in Acute Pancreatitis. Mol Med. 2014. 10.2119/molmed.2014.00117 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tang D, Kang R, Coyne CB, Zeh HJ, Lotze MT. PAMPs and DAMPs: signal 0s that spur autophagy and immunity. Immunological reviews. 2012;249(1):158–75. 10.1111/j.1600-065X.2012.01146.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Biswas SK, Mantovani A. Macrophage plasticity and interaction with lymphocyte subsets: cancer as a paradigm. Nat Immunol. 2010;11(10):889–96. 10.1038/ni.1937 . [DOI] [PubMed] [Google Scholar]
- 55.Gottschalk C, Mettke E, Kurts C. The Role of Invariant Natural Killer T Cells in Dendritic Cell Licensing, Cross-Priming, and Memory CD8(+) T Cell Generation. Front Immunol. 2015;6:379 10.3389/fimmu.2015.00379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Martinez LR, Mihu MR, Gacser A, Santambrogio L, Nosanchuk JD. Methamphetamine enhances histoplasmosis by immunosuppression of the host. J Infect Dis. 2009;200(1):131–41. 10.1086/599328 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mantri CK, Mantri JV, Pandhare J, Dash C. Methamphetamine inhibits HIV-1 replication in CD4+ T cells by modulating anti-HIV-1 miRNA expression. Am J Pathol. 2014;184(1):92–100. 10.1016/j.ajpath.2013.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jiang J, Wang M, Liang B, Shi Y, Su Q, Chen H, et al. In vivo effects of methamphetamine on HIV-1 replication: A population-based study. Drug Alcohol Depend. 2016;159:246–54. 10.1016/j.drugalcdep.2015.12.027 . [DOI] [PubMed] [Google Scholar]
- 59.Nair MP, Saiyed ZM, Nair N, Gandhi NH, Rodriguez JW, Boukli N, et al. Methamphetamine enhances HIV-1 infectivity in monocyte derived dendritic cells. J Neuroimmune Pharmacol. 2009;4(1):129–39. 10.1007/s11481-008-9128-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liang H, Wang X, Chen H, Song L, Ye L, Wang SH, et al. Methamphetamine enhances HIV infection of macrophages. Am J Pathol. 2008;172(6):1617–24. 10.2353/ajpath.2008.070971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Taylor HE, Simmons GE Jr., Mathews TP, Khatua AK, Popik W, Lindsley CW, et al. Phospholipase D1 Couples CD4+ T Cell Activation to c-Myc-Dependent Deoxyribonucleotide Pool Expansion and HIV-1 Replication. PLoS Pathog. 2015;11(5):e1004864 10.1371/journal.ppat.1004864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gauvin J, Chagnon-Choquet J, Poudrier J, Roger M, Montreal Primary HIVI, Slow Progressor C. Fluctuations in Blood Marginal Zone B-Cell Frequencies May Reflect Migratory Patterns Associated with HIV-1 Disease Progression Status. PLoS One. 2016;11(5):e0155868 10.1371/journal.pone.0155868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wilkinson D. Cofactors provide the entry keys. HIV-1. Curr Biol. 1996;6(9):1051–3. . [DOI] [PubMed] [Google Scholar]
- 64.Maruta T, Nihira M, Tomita Y. [Histopathological study on acute poisoning of methamphetamine, morphine or cocaine]. Nihon Arukoru Yakubutsu Igakkai Zasshi. 1997;32(2):122–38. . [PubMed] [Google Scholar]
- 65.Townley-Tilson WH, Callis TE, Wang D. MicroRNAs 1, 133, and 206: critical factors of skeletal and cardiac muscle development, function, and disease. Int J Biochem Cell Biol. 2010;42(8):1252–5. 10.1016/j.biocel.2009.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pankratz F, Hohnloser C, Bemtgen X, Jaenich C, Kreuzaler S, Hoefer I, et al. MicroRNA-100 Suppresses Chronic Vascular Inflammation by Stimulation of Endothelial Autophagy. Circ Res. 2018;122(3):417–32. 10.1161/CIRCRESAHA.117.311428 . [DOI] [PubMed] [Google Scholar]
- 67.Unlu S, Tang S, Wang E, Martinez I, Tang D, Bianchi ME, et al. Damage associated molecular pattern molecule-induced microRNAs (DAMPmiRs) in human peripheral blood mononuclear cells. PloS one. 2012;7(6):e38899 10.1371/journal.pone.0038899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xiao J, Tang J, Chen Q, Tang D, Liu M, Luo M, et al. miR-429 regulates alveolar macrophage inflammatory cytokine production and is involved in LPS-induced acute lung injury. Biochem J. 2015;471(2):281–91. 10.1042/BJ20131510 . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
This figure shows that rat body temperature is minimally affected by amphetamine (AMPH) when the room temperature is 16°C, but that severe hyperthermia usually occurs when AMPH is given and the room temperature is 23.5°C. It also shows that rats do not become hypothermic when given saline when the room temperature is 16°C.
(PDF)
This PCA plot shows that the Cool Saline (given saline in a 16°C rather than 23.5°C normal room temperature) animal transcriptome in blood was virtually identical to the normal room temperature saline animals. The red symbols for the Cool Saline Controls are embedded and obscured among the black symbols representing the Saline Controls (room temperature). It also shows the outlier in the Saline Controls and other groups that were removed from analysis and are not present in Fig 1A shown in the manuscript.
(PDF)
(DOCX)
(DOCX)
(DOCX)
(XLSX)
1st Sheet: Genes differentially expressed between AMPH normo and Control at FDR-adj-p < 0.05; 2nd Sheet: Genes differentially expressed between AMPH hyper and Control at FDR-adj-p < 0.05; 3rd Sheet: Genes differentially expressed between EIH and Control at FDR-adj-p < 0.05.
(XLSX)
1st Sheet: Genes differentially expressed between AMPH hyper and AMPH normo at FDR-adj-p < 0.05; 2nd Sheet: Genes differentially expressed between AMPH hyper and EIH at FDR-adj-p < 0.05.
(XLSX)
Only canonical pathways that had a p-value<0.01, │z-score│≥2. 0, and that contained at least three focus molecules for at least one of the treatment groups relative to control are listed.
(DOCX)
Data are presented as mean fold-change relative to control. *, p<0.05.
(DOCX)
(XLSX)
Data are presented as mean fold-change relative to control. * p<0.05.
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files. The RNA-seq data are available from NCBI GEO database (GSE62368, GSE64778, GSE115835).