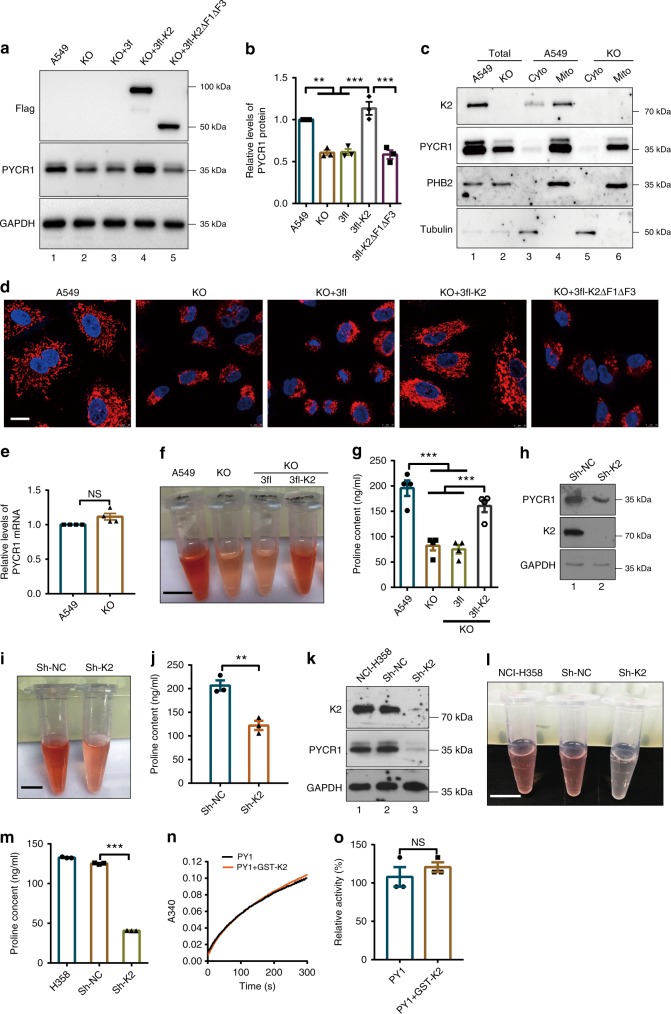
Fig. 4.
Kindlin-2 regulates PYCR1 and proline levels. a Kindlin-2 KO cells were infected with lentiviral vectors encoding 3xFLAG-tagged kindlin-2 (3fl-K2), 3xFLAG-tagged kindlin-2 F1, and F3 deletion mutant (3fl-K2∆F1∆F3) or 3xFLAG vector lacking kindlin-2 sequence (3fl). Three days later, the cells were analyzed by western blotting. b Protein levels of PYCR1 relative to that of GAPDH were quantified by densitometric analyses. The levels of PYCR1 in KO, 3fl, 3fl-K2, or 3fl-K2∆F1∆F3 cells were compared with that of A549 cells (normalized to 1) (n = 3). c The cytosolic (lane 3 and 5), mitochondrial (lane 4 and 6), and total (lane 1 and 2) lysates from A549 or kindlin-2 KO cells were analyzed by western blotting. d The cells were immunofluorescently stained with DAPI and anti-PYCR1 antibodies. Scale bar = 10 μm. e The mRNA levels of PYCR1 in kindlin-2 KO cells were analyzed by RT-PCR (n = 4) and compared with those in A549 cells (normalized to 1). f, g The proline levels were analyzed. The results were visualized (f) and quantified (g, n = 4). Scale bar = 1 cm. h–j A549 cells infected with kindlin-2 shRNA (Sh-K2) or control (Sh-NC) lentivirus were analyzed by western blotting (h). The proline levels were analyzed. The results were visualized (i) and quantified (j, n = 3). Scale bar = 1cm. k–m NCI-H358 cells were infected with kindlin-2 shRNA (Sh-K2) or control (Sh-NC) lentivirus for 5 days and then analyzed by western blotting (k). l, m The proline levels were analyzed. The results were visualized (l) and quantified (m, n = 3). Scale bar = 1 cm. n, o The effect of kindlin-2 on PYCR1 enzyme activity. The enzyme activity of purified PYCR1 (PY1) with or without GST-kindlin-2 (GST-K2) was analyzed. n The absorbance at 340 nm during the first 5 min of the reaction. o The relative activities of PYCR1 with or without GST-Kindlin-2 were calculated from n (n = 3). Data in b, e, g, j, m, and o represent means ± SEM. Statistical significance was calculated using one-way ANOVA with Tukey–Kramer post-hoc analysis in b, j, and m or using two-tailed unpaired Student’s t-test in e, j, and o, **P < 0.01; ***p < 0.001. NS, no significance