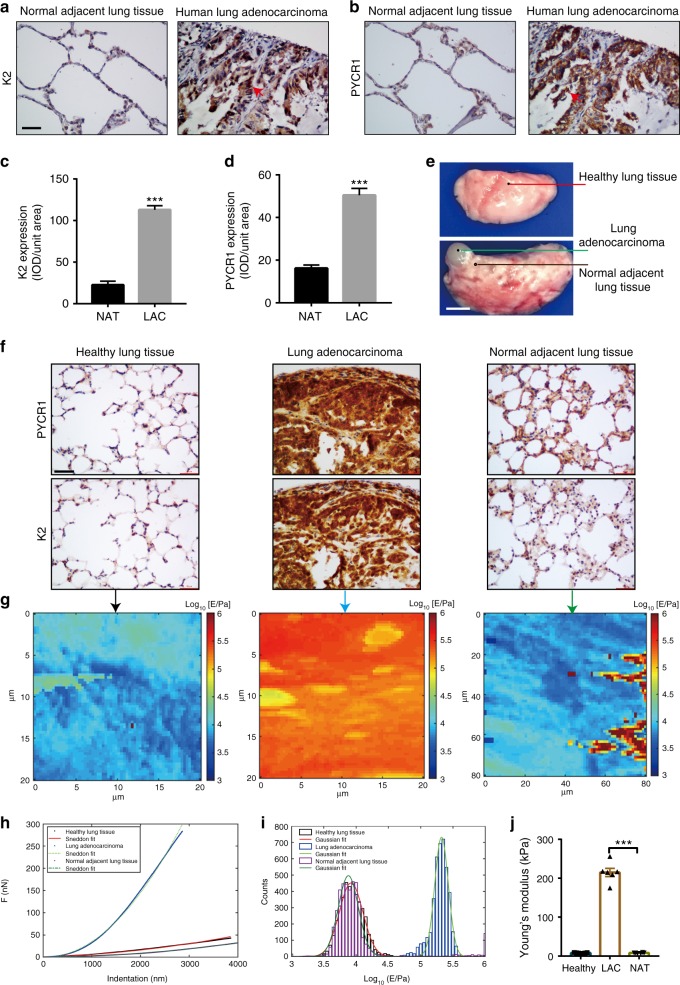
Fig. 8.
Elevated levels of kindlin-2, PYCR1, and tissue stiffness in lung adenocarcinoma. a–d Human lung adenocarcinoma (LAC) and normal adjacent lung tissues (NAT) were stained with kindlin-2 (a) or PYCR1 (b) antibodies as indicated. Red arrows indicate tumor regions. Scale bar, 20 μm. Kindlin-2 (c) and PYCR1 (d) immunohistochemical (IHC) staining of human lung adenocarcinoma and normal adjacent lung tissues was quantified as described in the Methods. Lung adenocarcinoma, n = 24; normal adjacent lung tissue, n = 8. ***P < 0.001. e–j Lung adenocarcinoma was induced by administration of Ad-Cre into the lung of Kras LSL−G12D/+ mice. Sixteen weeks later, lung tissues from the Kras LSL−G12D/+ mice administrated with Ad-Cre (e, lower panel) or without Ad-Cre as a control (e, upper panel) were shown. Scale bar, 500 μm. f Sections from areas of the tissues shown in e (as indicated in the figure) were analyzed by immunostaining with anti-PYCR1 and kindlin-2 antibody (f) and atomic force microscopy (g–j). Scale bar in f = 20 μm. Stiffness mapping of tissues are shown in g; force (nN) vs. indentation depth (nm) graph highlighting the raw data and fitting by the Sneddon model to extract the tissues’ elastic moduli (h). Quantitative analysis using histograms of Young’s modulus values in log-normal scale with a Gaussian distribution fit (i) and Young’s moduli (j, n = 6 distinct locations that are macroscopically distant from each other) of the health lung tissue, lung adenocarcinoma (LAC), and normal tissue adjacent to the tumor (NAT) were analyzed as described in the Methods. Data in c, d, j are presented as mean ± SEM using one-way ANOVA with Tukey–Kramer post-hoc analysis (j) or two-tailed unpaired Student’s t-test (c, d), ***P < 0.001