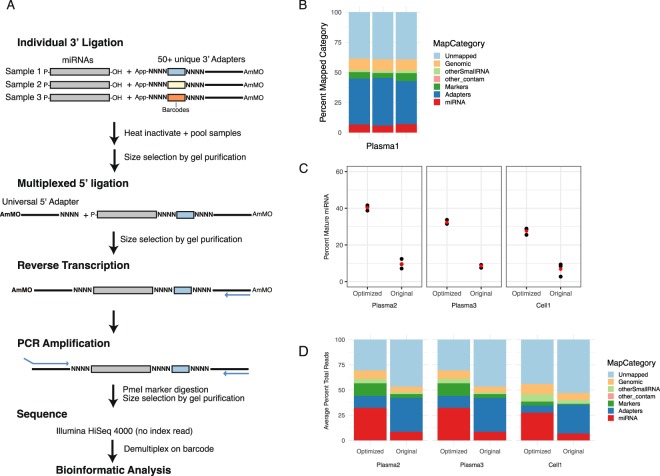
Figure 1.
Modifications to high-throughput sequencing method improves capture of miRNAs. (A) Schematic of protocol to prepare miRNA libraries for sequencing. Modifications from original protocol noted in bold. (B) Percentage of different classes of RNAs captured from a plasma sample using the original conditions (0.85 μM 3′ adapter, 3.3 μM unmodified 5′ adapter). Ligations were performed in triplicate from the same RNA. Each replicate is shown as an individual bar. Note the low percentage of reads mapping to miRNAs (red). (C) Percentage of mature miRNAs captured using the optimized conditions (0.05 μΜ 3′ adapter, 0.33 μΜ amino-modified 5′ adapter) were compared to original conditions in three independent biological samples. The ligation reactions were performed in triplicate for each sample and protocol. Each replicate is shown as a black dot. Red dot represents average. (D) The average percentage of reads mapping to different classes of RNA for each sample and condition shown in C.