Figure 5.
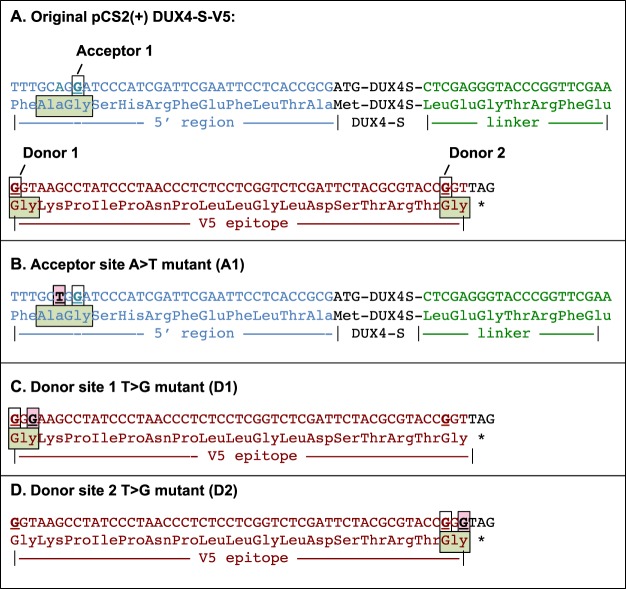
Acceptor and donor splice sites in the pCS2(+)-based DUX4-S-V5 plasmid and generation of mutant plasmids used to test functionality of these sites. (A) Based on sequencing of bands “B,” “D,” and “F” in Fig. 3C, we identified, as indicated, an acceptor site (blue G, boxed and underlined) in the 5’ region upstream of the DUX4-S cDNA. This acceptor sequence had a NetGene2 acceptor prediction confidence = 0.71. The sequencing also identified a first donor site (Donor 1, brown G, boxed and underlined), which had a NetGene2 splice donor site prediction confidence = 0.79. A second possible donor site (Donor 2, brown G, boxed and underlined) was located at the downstream (3′) end of the V5 epitope sequence as noted by Ansseau et al.23. This site had a NetGene2 splice donor site prediction confidence = 0.62. The amino acid(s) encoded near the acceptor and donor splice sites are shown in light green boxes. (B) To disable the acceptor site, we produced the A1 mutant by making an A to T switch of the nucleotide outlined by the light red box in the 5′ region upstream of the DUX4-S cDNA. (C) To disable the donor 1 site, we produced the D1 mutant by making a T to G switch of the nucleotide outlined by the light red box near the 5′ end of the V5 epitope. (D) To disable the donor 2 site, we produced the D2 mutant by making a T to G switch of the nucleotide outlined by the light red box at the 3′ end of the V5 epitope. Note that the encoded amino acid sequence was not altered by any of the three mutations. In addition, NetGene2 analyses predicted that none of the mutated sequences would function as acceptors or donors.