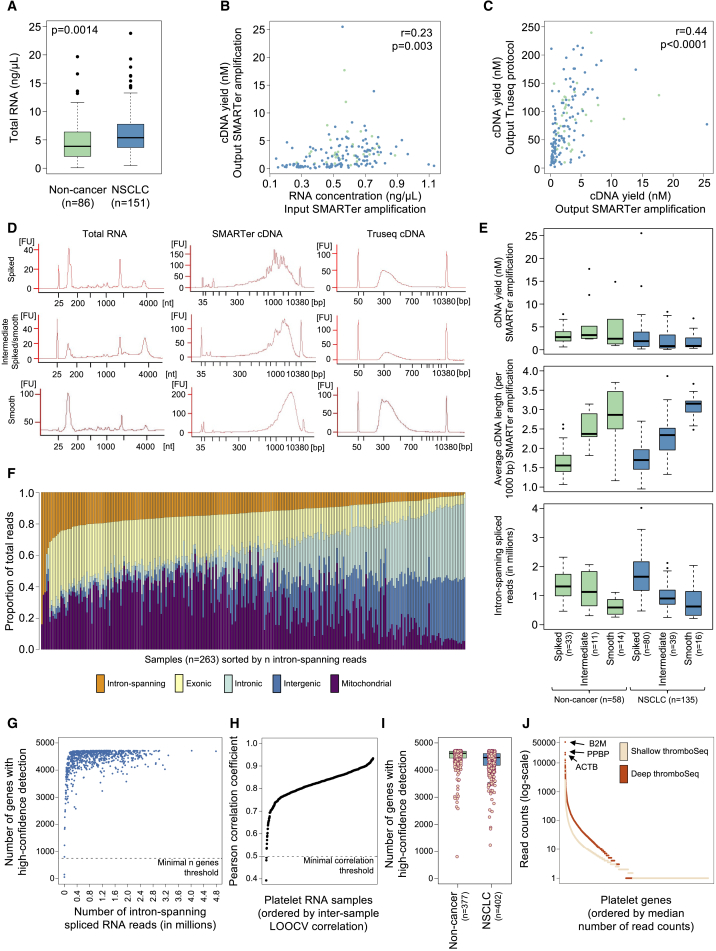
Figure 1.
Technical Performance of thromboSeq
(A) Platelet total RNA yield in ng/μL isolated from 6 mL whole blood in EDTA-coated Vacutainers tubes. p Value calculated by independent Student's t test.
(B) Correlation plot (Pearson's correlation) of estimated RNA input to the output SMARTer cDNA yield. Each dot represents a sample, color-coded by clinical group.
(C) Correlation plot (Pearson's correlation) of SMARTer cDNA yield to the Truseq cDNA library yield. Each dot represents a sample, color-coded by clinical group.
(D) Bioanalyzer profiles of samples with spiked, smooth, and intermediate spiked/smooth traces for total RNA as measured by RNA 6000 Picochip (left column), SMARTer amplified cDNA as measured by DNA High Sensitivity chip (middle column), and Truseq cDNA libraries as measured by DNA 7500 chip (right column).
(E) Shown are for spiked, smooth, and intermediate spiked/smooth SMARTer cDNA trace profiles for both non-cancer and NSCLC are the relative cDNA yield in nmol following SMARTer amplification (top), average cDNA length per 1,000 bp following SMARTer amplification (middle), and the number of intron-spanning spliced RNA reads (bottom).
(F) Selection of intron-spanning spliced RNA reads for thromboSeq analysis. Stackplot indicates the distribution of reads for each sample, subspecified from intron-spanning (orange), exonic (yellow), intronic (green), intergenic (blue), and mitochondrial (purple) regions.
(G) Selection of samples with >750 genes detected for thromboSeq analysis.
(H) Leave-one-sample-out cross-correlation filtering step for which counts of each sample were correlated to the median counts of all other samples.
(I) Number of genes detected with confidence in the platelet RNA samples using shallow thromboSeq.
(J) Deep thromboSeq versus shallow thromboSeq for matched platelet samples. Median total read counts (min-max) in millions: 59.7 (43.2–96.2) for deep thromboSeq and 12.9 (11.6–20.0) for shallow thromboSeq. The three genes with highest expression in deep thromboSeq are highlighted. The boxes of the boxplots indicate the interquartile range (IQR), the horizontal black line indicates the median values, and the whiskers range 1.5× the IQR. See also Figure S1 and Table S1.