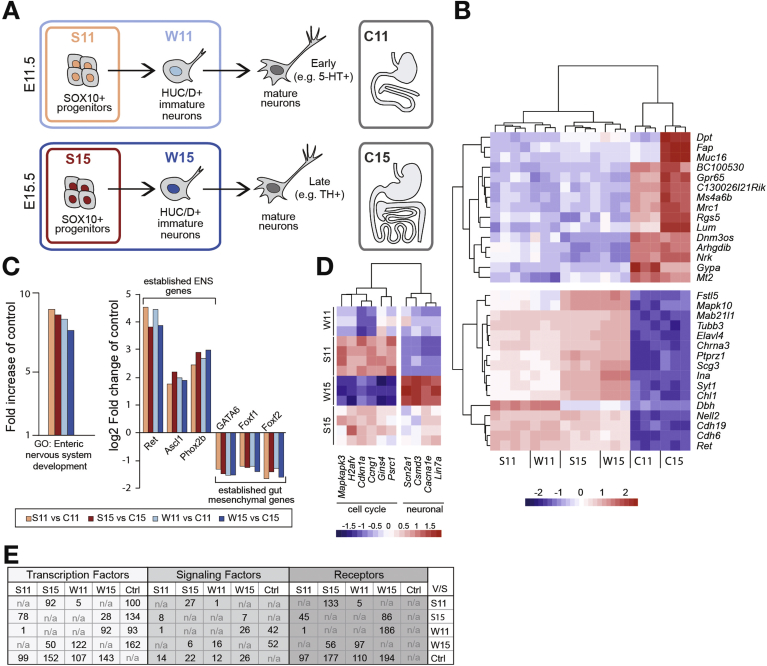
Figure 1.
Transcriptome screen design, verification, and identification of enriched genes. (A) Schematic drawing depicting the transcriptome analysis. ENS populations included SOX10+ progenitors (red boxes) and the whole ENS (blue boxes), each at 2 distinct development stages: E11.5 (S11 and W11) and E15.5 (S15 and W15). W11 contained immature neurons differentiating into, for example, 5-HT+ neurons, whereas W15 included immature neurons differentiating into other subtypes (eg, TH+). Non-ENS control gut tissue (gray boxes) at E11.5 (C11) and E15.5 (C15) was also included. (B) Heat map summarizing differentially expressed genes, compiled from the union of genes with top-10 absolute fold change (and P < .05) in the comparisons of S11vsC11, S15vsC15, W11vsC11, and W15vsC15. Genes and conditions are clustered by their hierarchical similarity. Color intensity represents the mean-centred log2 expression values. (C) Graphs comparing gene ontology (GO) term enrichment, ENS, and gut marker genes in the 4 ENS populations to control populations. (D) Heat map depicting cell cycle or neuronal genes. Genes are clustered by their hierarchical similarity. Color intensity represents the mean-centred log2 expression values. (E) Tables showing the number of transcription factors, signaling factors, and receptors found in pairwise comparisons (absolute fold change >1.2; P < .05) of the transcriptomes. n/a, not analyzed.