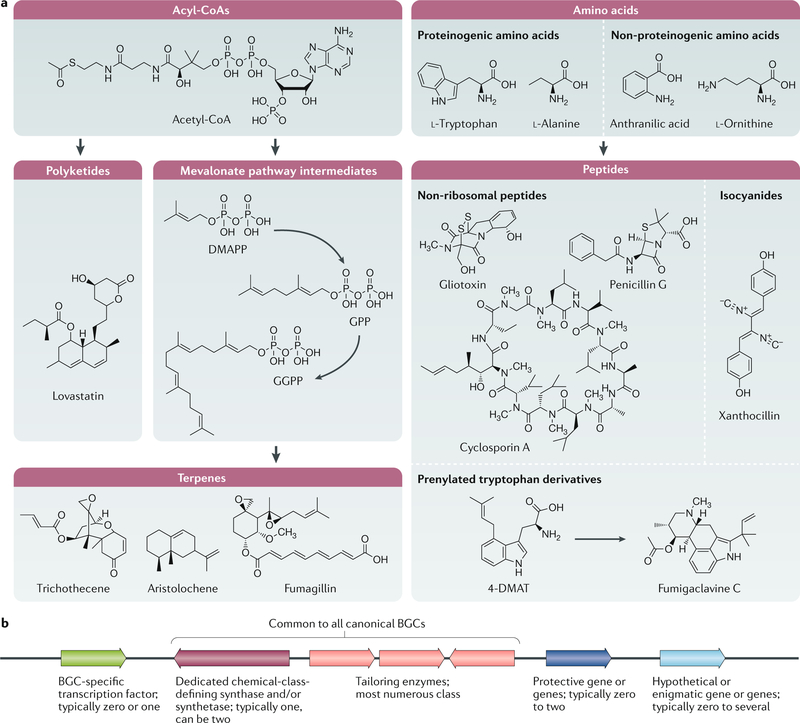
Fig. 1 |. The typical building blocks of secondary metabolites and a schematic overview of a biosynthetic gene cluster.
a | Most secondary metabolites can be grouped into three chemical categories: polyketides derived from acylCoAs, terpenes derived from acyl-CoAs and small peptides derived from amino acids. Hybrid molecules (polyketide– terpene, non-ribosomal peptide–polyketides and polyketide–fatty acid) are not shown. Fatty acid synthases (not shown) can occasionally contribute to the biosynthesis of secondary metabolites (for example, aflatoxin and sterigmatocystin are polyketide–fatty acid hybrids). b | Biosynthetic gene clusters (BGCs) are minimally composed of a chemically defining synthase and/or synthetase (polyketide synthase, terpene synthase and/or cyclase, non-ribosomal synthetase and isocyanide synthase) that use primary metabolites to form carbon backbones that are further modified by tailoring enzymes (for example, methyltransferases, p450 monooxygenases, hydroxylases and epimerases). Some BGCs contain cluster-specific transcription factors that typically positively regulate the other genes within the BGC; genes that encode proteins that mitigate the toxic property of the BGC secondary metabolite; and incongruous genes with hypothetical functions that are not obviously involved in the production of secondary metabolites or protection from the encoded metabolite. DMAPP, dimethylallyl diphosphate; DMAT, dimethylallyl tryptophan; GGPP, geranylgeranyl diphosphate; GPP, geranyl diphosphate. Part a adapted with permission from reF.30, Springer Nature Limited.