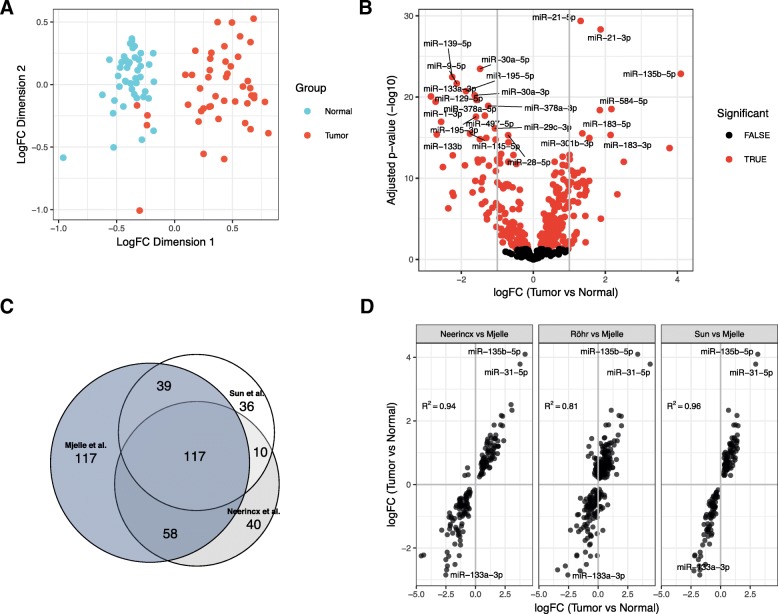
Fig. 1.
MicroRNAs and isomiRs are consistently differentially expressed between tumors and normal colon. a Multidimensional scaling plot (MDS plot) of the canonical miRNAs. Tumor samples are depicted in turquoise and matched adjacent normal samples are depicted in red. b Volcano plot showing expression differences for individual miRNAs between tumor and normal. The x-axis shows fold change values (log2) between tumor and normal tissue. The statistical comparison used was tumor vs normal such that a positive fold change indicates that the miRNA is upregulated in tumor tissue compared to normal tissue. The y-axis shows the minus log10 adjusted p-value, such that a higher value corresponds to higher significance. Red dots indicate that the miRNA is significant with adjusted p-value less than 0.05. c Venn-diagram showing the number and overlap of significant miRNAs in our dataset and the datasets of Neerincx and Sun. d Scatterplot comparing fold-change values between datasets. The most significantly upregulated miRNAs are shown with names as well as miR-133 which is the miRNA with the lowest logFC in our data. The statistical comparison is described in B). The correlation value is the Pearson correlation between the logFC values in the two datasets that are being compared