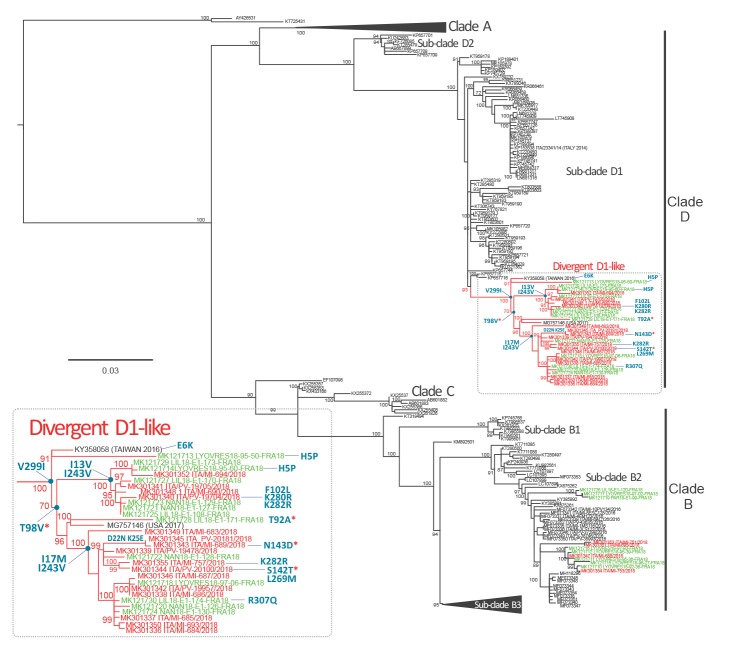
Figure.
Phylogenetic rooted tree inferred by maximum likelihood of complete enterovirus D68 VP1 sequences, northern Italy, 1 August–30 October 2018 (n = 20)
EV-D68: enterovirus D68; VP: viral protein.
A Tamura 3-parameter substitution model with a gamma distribution among-site rate variation was selected as the best-fit model by ModelTest in MEGA software 7.0.21 [21]. Significant branch support values (>70%) are given for maximum-likelihood implementations (IQ-TREE bootstrap support) and are reported near the branch nodes. The EV-D68 strains identified in this study are reported in red. EV-D68 strains identified in France in 2018 [22] are reported in green. Amino acids residues characterising divergent D1-like strains are indicated in blue, including mutations in the BC and DE loops reported with an asterisk.