Figure 6.
Proteasome Fragments αS Fibrils into Cytotoxic Species
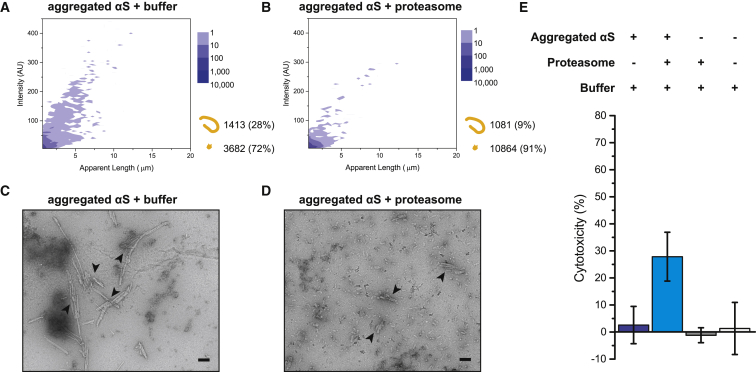
(A and B) Aggregates were assembled from αS monomers for 24 h and subsequently incubated with (A) an ATP-containing buffer or (B) proteasome holoenzyme for 20 h and presented in 2D plots as in Figure 2. Combined data of three independent repeats are shown (n = 3).
(C and D) The assay was repeated with untreated (C) or proteasome-treated (D) aggregated αS for TEM imaging. Arrows indicate typical aggregated structures. The scale bars represent 100 nm.
(E) HEK293 cells incubated with untreated (column 1) and proteasome-treated (column 2) samples of aggregated αS proteins, the buffer (column 3), or the proteasome alone (column 4) were tested for cytotoxicity using the LDH assay as in Figure 5A. Mean values are shown, with error bars representing SD.
All experiments were independently repeated at least three times (n = 3) using fresh αS and proteasome preparations.