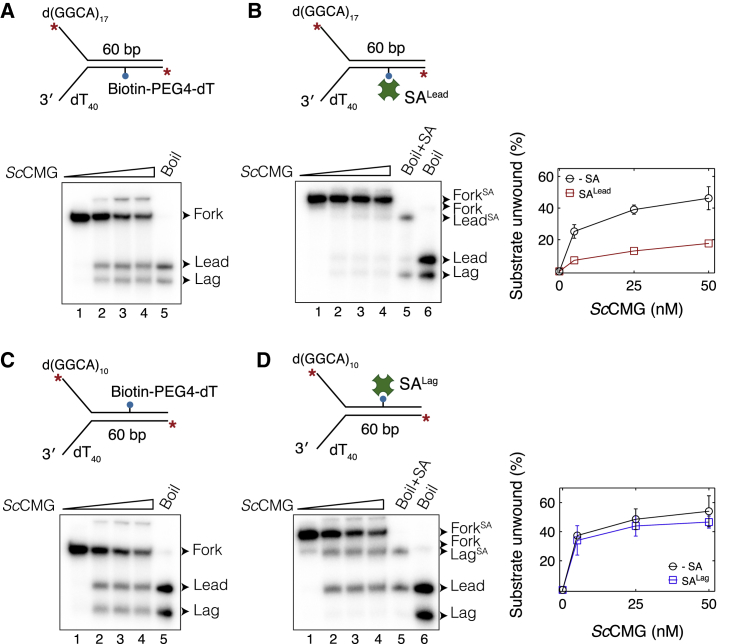
Figure 2.
ScCMG Bypasses a Biotin-Streptavidin Complex on the Excluded Strand
(A and B) Unwinding of fork DNA by ScCMG in the (A) absence or (B) presence of SALead. Right panel shows percentage of substrate unwound against ScCMG concentration.
(C and D) Unwinding of fork DNA by ScCMG in the (C) absence or (D) presence of SALag. Right panel shows percentage of substrate unwound against ScCMG concentration. In all gel images, lanes 1–4 correspond to reactions with 0, 5, 25, and 50 nM ScCMG. In panels (B) and (D), heat-denatured fork DNA was incubated with SA (lane 5) revealing the positions of SA-bound leading- (LeadSA) and lagging-strand (LagSA) templates. All fork DNA templates used in these assays were labeled at both 5′ ends with 32P. The radiolabel is shown as a red asterisk. Data represented here are mean ± SD from three independent experiments.
See also Figure S2.