Figure 6.
Single-Molecule Detection of CMG Pausing at a Lagging-Strand Methyltransferase Block
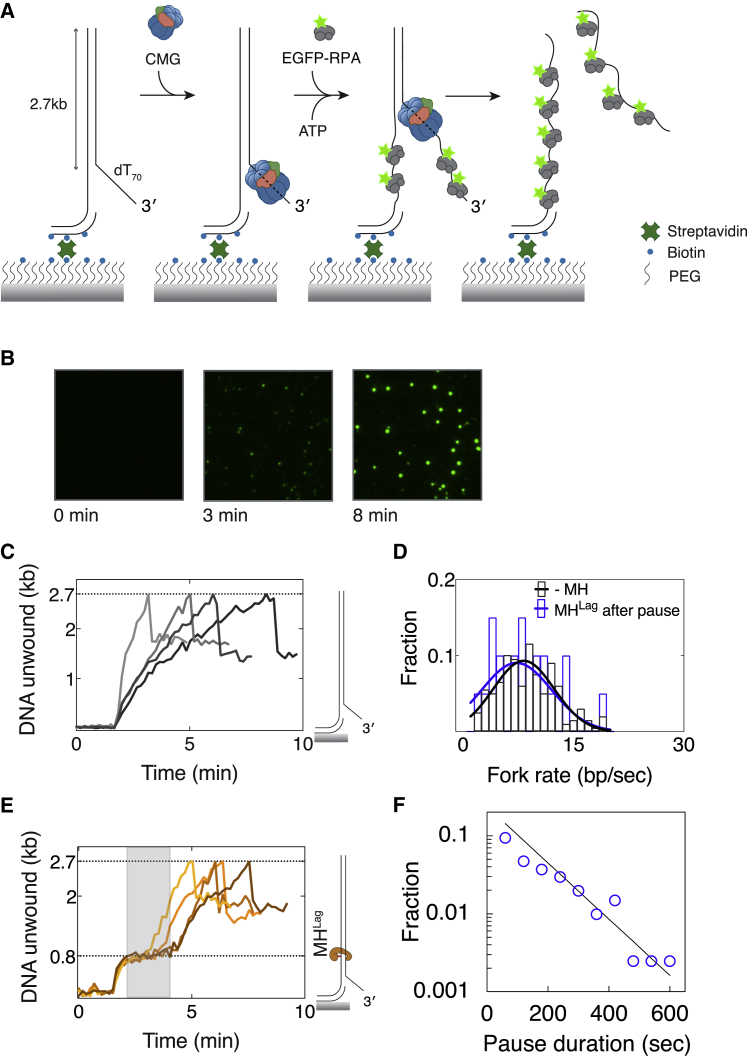
(A) Schematic representation of experimental approach used in single-molecule DNA unwinding assays.
(B) Images of a sample field of view showing accumulation of EGFP-RPA fluorescence signal at different time points from the addition of EGFP-RPA into the chamber.
(C) Example unwinding traces of DNA substrates without a protein barrier. Traces exhibit a signal drop upon completion of unwinding due to dissociation of the leading-strand template (depicted in A).
(D) Distribution of average fork rates measured in fully unwound substrates without MH (black) and after bypassing MHLag (blue). Number of molecules are n(-MH) = 199, n(MHLag after pause) = 20.
(E) Sample unwinding traces of DNA substrates modified with MHLag. Pausing observed at 800 bp is highlighted with gray rectangle.
(F) Distribution of pause durations observed in molecules exhibiting a pausing event (n = 109). The solid line is a fit to a single exponential.
See also Figure S6.