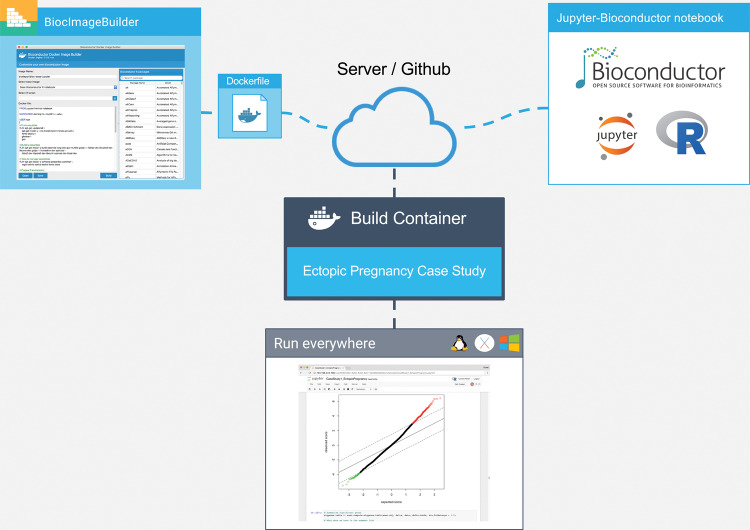
Figure 1.
Overview of our approach. The author of the Bioconductor workflow uses BiocImageBuilder to generate a Dockerfile that describes the Bioconductor and CRAN packages installed. The Dockerfile and the notebook files are uploaded to a server or GitHub repository. A custom container is then built with the default Linux base image for Bioconductor, dependencies for Jupyter, JuptyerHub, and/or Binder, and the Bioconductor packages. For GitHub installations, the Binder server builds the container and provides a link to run the container on its public cluster. JupyterHub provides the same functionality locally or on a private server. Using the container, the end user is able to view the notebook and execute, modify, and save the code on his or her local machine regardless of whether it uses Linux, MacOS, or Windows. In the case where the container is run remotely, no additional installation of software is required on the part of the end user.