Abstract
Background: Breast cancer patients at high risk for recurrence are treated with anthracycline-based chemotherapy, but not all patients do equally benefit from such a regimen. To further improve therapy decision-making, biomarkers predicting outcome are of high unmet medical need. Methods: The percent DNA methylation ratio (PMR) of the promoter gene coding for the Paired-like homeodomain transcription factor 2 (PITX2) was determined by a validated methylation-specific real-time polymerase chain reaction (PCR) test. The multicenter study was conducted in routinely collected archived formalin-fixed paraffin-embedded (FFPE) tissue from 205 lymph node-positive breast cancer patients treated with adjuvant anthracycline-based chemotherapy. Results: The cut-off for the PITX2 methylation status (PMR = 12) was confirmed in a randomly selected cohort (n = 60) and validated (n = 145) prospectively with disease-free survival (DFS) at the 10-year follow-up. DFS was significantly different between the PMR ≤ 12 versus the PMR > 12 group with a hazard ratio (HR) of 2.74 (p < 0.001) in the validation cohort and also for the patient subgroup treated additionally with endocrine therapy (HR 2.47; p = 0.001). Conclusions: Early-stage lymph node-positive breast cancer patients with low PITX2 methylation do benefit from adjuvant anthracycline-based chemotherapy. Patients with a high PITX2 DNA methylation ratio, approximately 30%, show poor outcome and should thus be considered for alternative chemotherapy regimens.
Key Words: PITX2 DNA methylation, Therapy prediction, Breast cancer
Introduction
Anthracycline-based chemotherapy has proven efficacy as adjuvant treatment in early breast cancer patients with lymph node-positive disease [1]. However, controversial discussions have emerged about the possibility of long-term anthracycline-related toxic effects, particularly cardiotoxicity like congestive cardiac failure and the risk of acute myeloid leukemia or myelodysplastic syndrome [2]. According to current national and international guidelines, standard of care are anthracyclines followed by taxanes. Also taxanes have significant toxic side effects, especially polyneuropathy. It has been shown that anthracycline-free regimens are not inferior to anthracyclines [3]. The question arises whether patients require both chemotherapy components and whether some patients might be sufficiently treated with anthracycline or taxanes alone. Subsequently, a significant number of breast cancer patients could be spared to experience such severe secondary diseases if predictive tests were available to identify patients with good outcome after anthracycline-based chemotherapy or taxane alone and thus would support the treating physicians to apply such therapy. Such a fundamental improvement of adjuvant chemotherapy selection would require predictive tests for both anthracyclines and taxanes.
Paired-like homeodomain transcription factor 2 (PITX2) functions in the Wnt signaling pathway by recruiting and stabilizing cytosolic β-catenin, which triggers transcription of cell cycle-regulatory and proliferation genes (e.g. those coding for cyclin D1 and c-Myc) and subsequently enhances cell proliferation [4]. Methylation of the PITX2 gene promoter leads to epigenetic silencing of this transcription factor and subsequent changes in molecular signaling. DNA methylation plays a crucial role in the development and progression of a variety of cancers, including breast cancer [5,6,7,8,9].
Significant evidence has accumulated that methylation of the PITX2 gene promoter might serve as a predictive and prognostic biomarker in a variety of cancers, including high-risk breast cancer patients [10,11,12,13,14].
Hayes [15] meticulously defined the criteria for biomarker tests in order to qualify for clinical use: demonstration of analytical validity, proven clinical/biological validity and clinical utility. The present study describes the clinical utility of a PITX2 DNA methylation assay targeting the P2 promoter of the gene. This assay has been developed as an accurate, reproducible and reliable quantitative methylation-specific polymerase chain reaction (PCR) assay utilizing genomic DNA extracted from routinely available formalin-fixed primary breast cancer tumor tissue, thus fulfilling the requirements for analytical validity. Different methods of determining the PITX2 percent DNA methylation ratio (PMR) by microarray testing or PCR-based quantification resulted in comparable statistically significant PMR values to predict a patient's outcome while receiving adjuvant anthracycline containing therapy [10,14]. However, these assays were all research tests, not analytically validated and therefore do not qualify according to Hayes [15] as suitable for the clinical routine setting. The present study aimed at validating the PITX2 DNA methylation status by applying a fully analytically validated assay [16] in a prospective, predefined analysis to predict outcome in lymph node-positive breast cancer patients receiving adjuvant anthracycline-based chemotherapy. The results show that the PITX2 DNA methylation assay can be used to further improve the concept of individualized cancer therapy decision-making and can support physicians on a case-by-case basis to optimize the treatment plan for each single breast cancer patient.
Material and Methods
Patients, Samples, Study Design and Conduct
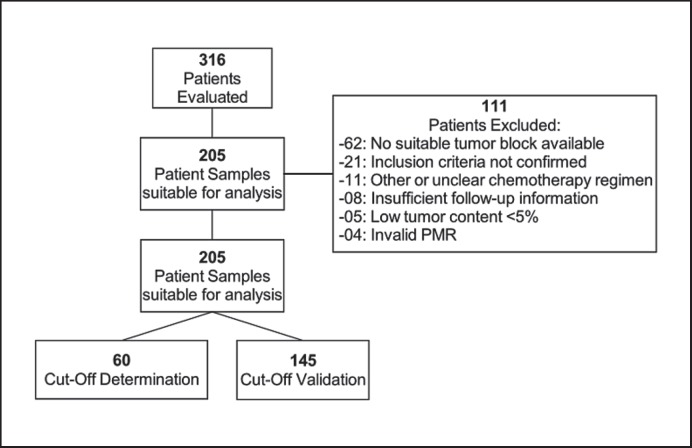
From 316 patients enrolled in this study who underwent surgery between 1987 and 2011, 205 patients were suitable for the final analyses with the following clinical and technical eligibility criteria: histologically confirmed invasive breast cancer, primary tumor stage pT1, pT2 or pT3, histologically confirmed lymph node involvement (≥N1), estrogen receptor(ER)-positive (progesterone positivity alone was not eligible), human epidermal growth factor receptor 2 (HER2)-negative (triple-negative breast cancers were excluded), standard-of-care adjuvant anthracycline-based chemotherapy (none of the patients received neoadjuvant treatment), no dose-dense therapy, no other primary systemic chemotherapy except for endocrine therapy, no additional taxanes, availability of clinical follow-up data for at least 18 months, valid PITX2 PMR values per tissue sample assessed. The Consolidated Standards of Reporting Trials (CONSORT) diagram (fig. 1) lists all patients assessed for eligibility and the reasons for exclusion.
Fig. 1.
CONSORT diagram. The diagram depicts the eligibility of patients and the respective samples that were included in the clinical validation study and the reasons for non-inclusion.
Surgical specimens from primary tumors (formalin-fixed paraffin-embedded (FFPE) tissue) and corresponding clinical data were obtained from the following 3 institutes: Klinikum rechts der Isar of the Technical University of Munich (TUM), Munich, Germany; Erasmus University Medical Center, Rotterdam, The Netherlands; Martin-Luther-University Halle, Halle (Saale), Germany. For all patients, source data verification was performed. Any experiments on humans cited were conducted in accordance with the Declaration of Helsinki (1964). Ethical committee approval or appropriate consent according to national law and institutional requirements are archived at each site.
A sample size of at least 65 events for the validation set study group was calculated based on available clinical data and a hazard ratio (HR) of 2.0 with a statistical significance level of α = 5% and a power of 80%. Based on these assumptions, a sample size of 145 patients (validation set study group) for the clinical validation study was determined. Therefore, out of the 205 patients, a training set of 60 patients was randomly selected (assuring the same percentage of patients experiencing an event and treatment with endocrine therapy) and used for determining the optimal PITX2 PMR cut-off value (training set cut-off study group). After confirmation of the clinical cut-off with the training set cut-off study group, the clinical validation study protocol with the predefined objectives of the study was locked. All procedures and above parameters for the prospective analysis were conducted as predefined in the study protocol. The characteristics of the patient populations are described in table 1.
Table 1.
Patient study groups and their characteristics
| Classification | Training set cut-off study group |
Validation set study group |
Clinical analysis study group |
|||
|---|---|---|---|---|---|---|
| n = 60 | % | n = 145 | % | n = 205 | % | |
| Age, years | ||||||
| <50 | 31 | 51.7 | 57 | 39.3 | 88 | 42.9 |
| ≥50 | 29 | 48.3 | 88 | 60.7 | 117 | 57.1 |
| T stage | ||||||
| T1 | 24 | 40.0 | 75 | 51.7 | 99 | 48.3 |
| T2, T3, T4, Tx | 36 | 60.0 | 70 | 48.3 | 106 | 51.7 |
| N stage | ||||||
| N1 | 45 | 75.0 | 110 | 75.9 | 155 | 75.6 |
| N2 | 13 | 21.7 | 34 | 23.4 | 47 | 22.9 |
| N3 | 2 | 3.3 | 1 | 0.7 | 3 | 1.5 |
| Grading G | ||||||
| G1 | 0 | 0.0 | 9 | 6.2 | 9 | 4.4 |
| G2 | 38 | 63.3 | 100 | 69.0 | 138 | 67.3 |
| G3 | 22 | 36.7 | 36 | 24.8 | 58 | 28.3 |
| Lymph nodes involved, n | ||||||
| 1–3 | 37 | 61.7 | 94 | 64.8 | 131 | 63.9 |
| >3 | 23 | 38.3 | 51 | 35.2 | 74 | 36.1 |
| ER positivity | 60 | 100 | 145 | 100 | 205 | 100 |
| HER2 negativity | 60 | 100 | 145 | 100 | 205 | 100 |
| Endocrine therapy | ||||||
| Yes | 41 | 68.3 | 99 | 68.3 | 140 | 68.3 |
| No | 19 | 31.7 | 46 | 31.7 | 65 | 31.7 |
| Disease recurrence | ||||||
| Yes | 31 | 51.7 | 74 | 51.0 | 105 | 51.2 |
| No | 29 | 48.3 | 71 | 49.0 | 100 | 48.8 |
| Disease recurrence (plus ET) | ||||||
| Yes | 18 | 30.0 | 43 | 29.7 | 61 | 29.8 |
| No | 23 | 38.3 | 56 | 38.6 | 79 | 38.5 |
| Overall survival | ||||||
| Yes | 23 | 38.3 | 50 | 34.5 | 73 | 35.6 |
| No | 37 | 61.7 | 95 | 65.5 | 132 | 64.4 |
| Radiotherapy | ||||||
| Yes | 42 | 70.0 | 106 | 73.1 | 148 | 72.2 |
| No | 18 | 30.0 | 39 | 26.9 | 57 | 27.8 |
| PMR (cut-off) | ||||||
| ≤12 | 41 | 68.3 | 102 | 70.3 | 143 | 69.8 |
| >12 | 19 | 31.7 | 43 | 29.7 | 62 | 30.2 |
ER = Estrogen receptor, HER2 = human epidermal growth factor receptor 2, ET = endocrine therapy, PMR = percent DNA methylation ratio.
PITX2 DNA Methylation Assay
All primary FFPE breast cancer tissue samples were coded to ensure blinding of the operator while conducting the PITX2 DNA methylation assay. The PITX2 test (QIAGEN therascreen® PITX2 RGQ PCR kit) is a quantitative methylation-specific real-time PCR test (qMSP), intended for the determination of the PMR in the promotor 2 (P2) of the PITX2 gene in primary FFPE breast cancer tissue [11]. Genomic DNA (gDNA) was extracted and purified from FFPE primary tumor tissue using the QIAamp DSP DNA FFPE Tissue Kit (QIAGEN, Hilden, Germany). After bisulfite conversion with the Epitect Fast DNA Bisulfite Kit (QIAGEN, Hilden, Germany), the PMR of 3 CpG motifs of the PITX2 gene P2 [11] was quantified by qMSP using the therascreen PITX2 RGQ PCR kit, containing the quantitative PCR (qPCR) reaction mix, primer and probes as well as positive and negative controls. The assay was performed on the Rotor-Gene Q MDx real-time PCR platform (QIAGEN, Hilden, Germany) and automatically assessed by the Rotor-Gene AssayManager® software V2.1 with installed Gamma Plug-in and PITX2-specific assay profile (therascreen_PITX2_FFPE_CE Assay Profile v1.0) for data analysis and quality control [16].
Statistical Methods
Disease-free survival (DFS) was the primary endpoint and defined as the time from primary surgery to the first documented DFS event. The date of primary surgery is considered as the follow-up index date. DFS events include reappearance of the cancer disease (local disease recurrence or distant metastasis), secondary malignancies considered life-threatening, and death of any cause. Secondary malignancy was observed in 6 cases: 2 patients with gastric cancer and 1 patient with acute lymphatic leukemia; these occurrences were classified as DFS events. 2 patients had malignancies with excellent prognosis, 1 of the corpus uteri and 1 of the bladder, and were not considered life-threatening cancers and therefore not classified as DFS events. 1 patient who died from colon cancer without recurrence of breast cancer was classified as death of any cause.
The PITX2 cut-off value for DFS was established with the ‘maximum-selected log-rank statistic’ using the maxstat.test function as implemented by the program library ‘maxstat’ of the program ‘R’ (R Development Core Team 2012) [17]. For patients (n = 15) who died without recurrence of cancer, competing risks analysis according to Fine and Gray [18] was applied. Analysis was performed for DFS follow-up time censored at 10 years. Survival curves were calculated according to the incidence function [18]. The primary objective was to confirm the predictive value of PITX2 DNA methylation applying the pre-defined cut-off value of PMR = 12 as stated in figure 1 in FFPE primary breast cancer tumor tissue samples in an independent patient cohort (n = 145). The study was considered successful if a statistically significant differentiation in the primary endpoint DFS was observed between patients with PMR ≤ 12 versus those with a PMR > 12. The log-rank test was used for calculating the respective p-values and a 2-sided significance level of 5% was considered significant. R software version: 3.4.1 (2017-06-30) - ‘Single Candle’, The R Foundation for Statistical Computing, 2017. The relationship between PITX2 DNA methylation and established clinical factors with the primary endpoint DFS was analyzed using univariate and multivariate Cox proportional hazard models [19,20].
Results
Patient Characteristics and Study Populations
Patient study groups and their characteristics are presented in table 1. All patients had received anthracycline-based chemotherapy consisting of epirubicin in combination with cyclophosphamide and fluorouracil or cyclophosphamide alone. Current national and international guidelines recommend anthracycline-taxane combination regiments. Patients treated with taxanes were not included in the present study because the primary goal was to validate the value of PITX2 DNA methylation to predict the outcome of patients receiving anthracyclines.
Noteworthy, the majority (68.3%) but not all patients (because the threshold of ER positivity changed during the enrollment period, >10% down to >1% stained tumor cells in order to qualify for ER positivity) received endocrine therapy (according to guidelines) in addition to chemotherapy. 15 patients died without experiencing disease recurrence, which prompted us to analyze the predictive value not only with Kaplan-Meier analysis but also with competing risks methodology [18]. The patient cohort (n = 205) was divided into a ‘training set cut-off study group’ (n = 60) and a ‘validation set study group’ (n = 145). The total patient cohort (‘clinical analysis study group’ n = 205) was used for additional clinical analyses.
PITX2 PMR Cut-off Confirmation
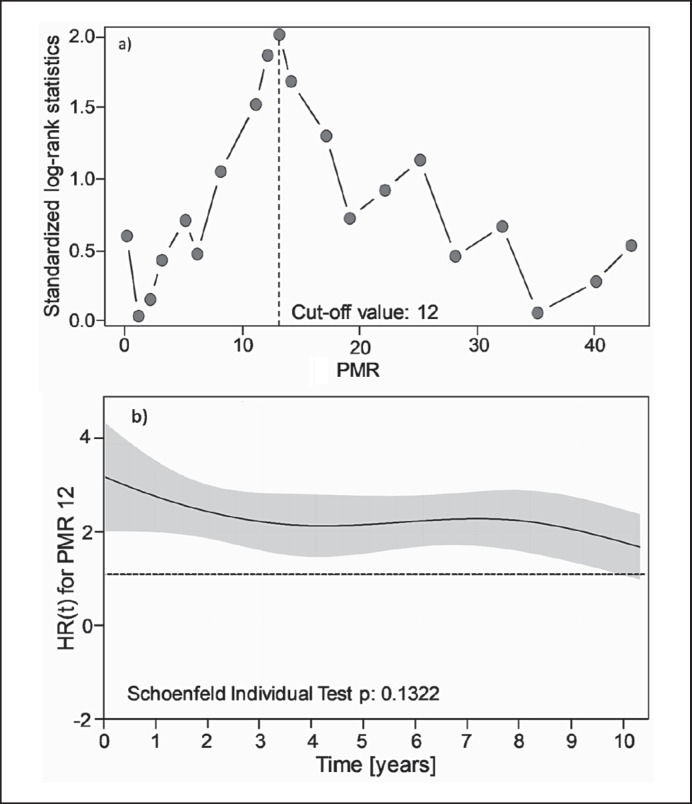
To determine a PMR cut-off that identifies patients with good and poor outcome, i.e. short and long DFS, original microarray data of a previous breast cancer study [10] were mathematically converted into PMR values. PITX2 DNA methylation determined by the PCR-based therascreen PITX2 RGQ PCR kit in 121 fresh-frozen tissue samples of that same study served as transformation curve to convert all data (n = 204) of that study [10] into PMR values. Applying log-rank statistics, a PITX2 cut-off value of PMR 12 was established (unpublished data). However, the previous studies used fresh-frozen tissue, and although Harbeck et al. [13] have demonstrated an excellent correlation for the PITX2 DNA methylation status analyzed by qPCR in fresh-frozen and FFPE tissue, we also reconfirmed, in a cohort of 60 breast cancer patients randomly selected from the clinical analysis study group (n = 205), the cut-off value in FFPE tissue (training set cut-off study group, patients treated with anthracycline-based chemotherapy including 41 patients treated additionally with endocrine therapy) (table 1). In these 60 patients, using gDNA extracted from primary breast cancer tumor FFPE tissue, the cut-off value of PMR = 12 was confirmed by log-rank statistics when tested over a PMR range from 0 to 50 (fig. 2a). Furthermore, applying the chi-squared test for the proportional hazards regression model (Schoenfeld residues plot; fig. 2b), the effect of the PITX2 DNA methylation cut-off at PMR 12 remained stable over the period of 10 years of follow-up time [21].
Fig. 2.
Cut-off determination and Schoenfeld plot. a Statistical determination of PITX2 DNA PMR cut-off points (n = 60); training set; competing risks analysis, 5-year time of follow-up. PMR data were determined with the therascreen® PITX2 RGQ PCR kit. y-Axis: standardized log-rank statistics; x-axis: PMR. The dashed vertical line marks the maximum log-rank statistic at a PITX2 PMR cut-off point of 12. b Schoenfeld residues plot of PITX2 PMR with respect to disease-free survival (10-year time of follow-up). Dashed line: hazard ratio (HR) of 1.0; solid line: HR applying the cut-off value of PMR 12 at a given time. The gray area represents the standard error.
Clinical Validation Study
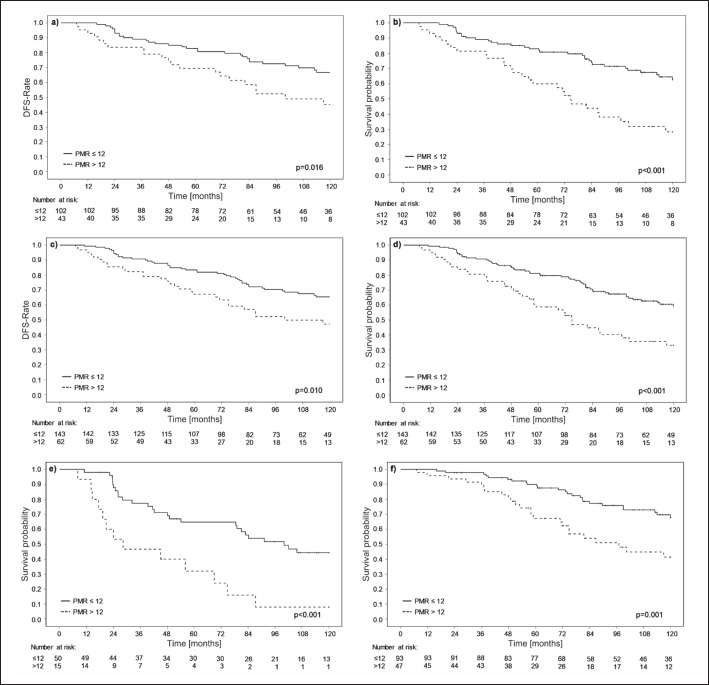
The clinical validation study (n = 145; validation set study group) was conducted under an approved study protocol with a predefined endpoint and a predetermined PITX2 DNA methylation PMR cut-off value of 12. The results demonstrate that patients with a PMR ≤ 12 (70.3% of patients) (table 1) do benefit from anthracycline-based chemotherapy while patients with PMR > 12 (29.7% of patients) (table 1) have a lower probability to benefit from this kind of chemotherapy (fig. 3).
Fig. 3.
Survival analysis. The high-risk breast cancer patient groups were subjected to competing risks analysis and Kaplan-Meier survival analysis with a follow-up time of 10 years. a Clinical validation set study group (n = 145); patients treated with anthracycline-based chemotherapy with and without endocrine therapy, competing risks analysis. b Clinical validation set study group (n = 145); patients treated with anthracycline-based chemotherapy with and without endocrine therapy, Kaplan-Meier analysis. c Clinical analysis study group (n = 205); patients treated with anthracycline-based chemotherapy with and without endocrine therapy, competing risks analysis. d Clinical analysis study group (n = 205), patients treated with anthracycline-based chemotherapy with and without endocrine therapy, Kaplan-Meier analysis. e Clinical analysis study group (n = 65); patients treated with anthracycline-based chemotherapy only, Kaplan-Meier analysis. f Clinical analysis study group (n = 140); patients treated with anthracycline-based chemotherapy plus endocrine therapy, Kaplan-Meier analysis.
In the validation set study group, 10 patients died (cause of death unknown) without experiencing recurrence from breast cancer. Survival analysis applying Kaplan-Meier analysis censors patients in case of death, implying these patients have the same risk as compared to the overall study patient population. In the situation where patients die without disease recurrence, the problem of competing risks arises. Therefore, we applied the competing risks method to reassure the significance of PITX2 DNA methylation. By applying the competing risks method, the PMR = 12 cut-off value divided the patients into 2 groups which had an HR for the primary endpoint of 1.97, which was statistically significant (p = 0.016; 95% confidence interval (CI) 1.14–3.43) (fig. 3a). Kaplan-Meier analysis resulted in HR = 2.74, with pronounced statistical significance (p < 0.001; 95% CI 1.65–3.54) (fig. 3b).
The clinical analysis study group (n = 205) was also subjected to competing risks and Kaplan-Meier analysis applying the cut-off of PMR 12. The competing risks analysis resulted in the separation of 2 patient populations with HR = 1.85 and p = 0.01 (95% CI 1.16–2.96) (fig. 3c) and the Kaplan-Meier analysis showed an even higher HR of 2.25 with high statistical significance (95% CI 1.47–3.44; p < 0.001) (fig. 3d). If secondary malignancies were not included as events in the statistical analysis, the value of PITX2 methylation was also highly statistical significant (n = 205; HR 1.85; p = 0.010), demonstrating no impact of secondary malignancies. The subgroup of patients who received anthracycline-based chemotherapy alone, also small in number, demonstrated in Kaplan-Meier analysis a clear and statistically significant distinction between 2 groups with an HR of 3.24 (95% CI 1.63–6.42; p < 0.001) (fig. 3e). By focusing on the patient population that has received endocrine therapy in addition to anthracycline-based chemotherapy within the clinical analysis study group, the predictive effect of PITX2 DNA methylation is pronounced; Kaplan-Meier analysis: HR = 2.47 with high statistical significance (95% CI 1.41–4.34; p = 0.001) (fig. 3f).
An important aspect for clinical decision-making is how many patients will be diagnosed with a PITX2 DNA methylation status that is close to the cut-off value. We analyzed how many patients will be clearly outside of a 20% window at the cut-off value of PMR 12. In our study, 90.7% of the patients had a PMR below 10 or above 14. Only 9.3% had PMR values within this artificial window. Therefore, the vast majority of patients will present with an unambiguous PITX2 DNA methylation status.
Multivariate Analysis
In multivariate Cox regression analyses in the validation set study group, the PITX2 DNA methylation status was found to contribute significant information regarding DFS (table 2). The cut-off of PMR = 12 demonstrates with an HR of 2.72 (95% CI 1.55–4.78) PITX2 DNA methylation as an independent factor with high statistical significance (p = 0.001) in multivariate analysis. Concerning the established prognostic factors, age and lymph node status (comparing less than 3 lymph nodes affected vs. more than 3 lymph nodes) did not contribute any additional information while pT stage did, but only if comparing pT1 versus pT2 and not pT1 versus pT3. The latter might be due to the low number of cases. Histological grading was not of statistical significance in the multivariate analysis. Besides the PITX2 DNA methylation status, also endocrine therapy contributed statistically significant information.
Table 2.
Multivariate Cox regression analysis to assess the clinical impact with respect to DFS of the PITX2 DNA methylation status to predict outcome of breast cancer patients after adjuvant anthracycline-based chemotherapy (validation set study group, n = 145)
| Variable | HR | 95% CI | p |
|---|---|---|---|
| T stage | |||
| T2 vs. T1 | 2.85 | 1.52–5.36 | 0.001 |
| T3 vs. T1 | 1.55 | 0.65–3.71 | 0.324 |
| Nodal status, LNs > 3 vs. 1–3 | 1.63 | 0.90–2.94 | 0.107 |
| Endocrine therapy, yes vs. no | 2.94 | 1.70–5.09 | 0.001 |
| Grading, G1/2 vs. G3 | 1.53 | 0.88–2.68 | 0.132 |
| Age, years, >50 vs. ≤50 | 1.22 | 0.71–2.09 | 0.462 |
| PITX2 DNA methylation status, | 2.72 | 1.55–4.78 | 0.001 |
| PMR ≤ 12 vs. PMR > 12 |
DFS = Disease-free survival, HR = hazard ratio, CI = confidence interval, LN = lymph node, PMR = percent DNA methylation ratio.
Relevance of Tumor Content in FFPE Tissue Sections
We observed a significant variation of tumor cell content in the tumor tissues assessed. In order to examine the PITX2 DNA methylation status dependency on the percentage of tumor cell content, the clinical analysis study group (n = 205) was divided into 2 populations, one with >50–90% tumor content (n = 89) and the other one with 5–50% tumor content (n = 116). Kaplan-Meier analysis employing the optimal cut-off value of PMR = 12 revealed very similar statistics: low tumor content: HR = 2.25 (95% CI 1.22–4.18) with a significance level of p = 0.010; high tumor content: HR = 2.08 (95% CI 1.18–3.66) with a significance of p = 0.012. Therefore, the PITX2 DNA methylation status can be determined by qPCR in gDNA extracted from primary breast cancer FFPE tissue independent from variation in tumor cell content, and macrodissection of tumor samples prior to analysis seems not to be required.
Discussion
Anthracyclines, typically in combination with taxanes, have provided compelling evidence for their efficacy in treating early breast cancer patients, both in the adjuvant and neoadjuvant setting [1]. National and international guidelines recommend anthracycline-based chemotherapy regimens as a most suitable treatment option [22,23], despite their considerable associated toxicity eventually causing leukemia or congestive heart failure. Unless non-anthracycline-based chemotherapy improves the outcome similar to anthracyclines, they will remain an integral part of adjuvant treatment to reduce the risk of disease recurrence as an alternative option, if treatment with anthracyclines is insufficient or ineffective. Predictive cancer biomarkers may help to decide if adjuvant systemic anthracycline-based chemotherapy is the right treatment option for early breast cancer patients. Our work shows that hypomethylation of the PITX2 gene promoter identifies a subgroup of lymph node-positive breast cancer patients treated with anthracycline-based chemotherapy with excellent outcome.
Several studies have addressed this so far unmet medical need for predictive biomarkers; however, none of those approaches have transferred into clinical routine. HER2 and topoisomerase IIA (TOP2A) have been evaluated for their use as predictive biomarkers for anthracycline responsiveness [24]. The authors came to the conclusion that both markers, either alone or in combination, have some added value in the prediction of outcome after anthracycline-based chemotherapy, but their findings do support their use only in patients with HER2-amplified or TOP2A-aberrant tumors. Desmedt et al. [25] developed an anthracycline-based score, the A-score, combining TOP2A and signatures of tumor invasion and immune response. This A-score demonstrated a high negative predictive value for anthracycline responsiveness, but additional validation, especially analytical validity, is missing so far. Commercially available gene signatures such as Endopredict® (Myriad Genetics Inc., Salt Lake City, UT, USA), OncotypeDX® (Genomic Health, Redwood City, CA, USA), or Mammaprint® (Agendia Inc., Irvine, CA, USA) have not provided evidence to be of merit to predict outcome after anthracyclines [23]. These multigene signatures are highly valuable to define patients who could be spared to receive chemotherapy and who are sufficiently treated with endocrine therapy alone [23].
The present retrospective study was conducted by applying a fully analytically validated quantitative methylation-specific real-time PCR format [16] and an optimized clinically validated cut-off to extend previously published studies. Hartmann et al. [10] investigated the value of PITX2 DNA methylation in 241 lymph node-positive, ER-positive and HER2-negative breast cancer patients, a patient cohort with the same clinicopathological characteristics as the present study, and showed the significant role of PITX2 DNA methylation in the prediction of outcome after adjuvant anthracycline treatment, at that time by applying a microarray-based test. Hartmann et al. [10] used arbitrary quartiles instead of an optimized cut-off, identified through log-rank statistics, to separate hypermethylated from hypomethylated tumor specimens, but they nevertheless demonstrated that patients with PITX2 hypermethylated tumors had a significantly worse outcome than patients with hypomethylated tumors. The hypothesis put forward to explain this effect is that hypermethylation of PITX2 results in silencing of the Wnt/β-catenin pathway with subsequent downregulation of estrogen and its receptor, causing estrogen deprivation/independency in estrogen-positive cancer cells. The ABCG2 transporter is downregulated by estrogen, i.e. estrogen deprivation leads to increased ABCG2 expression. If PITX2 is hypermethylated, the Wnt/β-catenin pathway is silenced and subsequent ER downregulation leads to overexpression of the ABCG2 transporter and thereby to anthracycline resistance [26,27].
Absmeier et al. [14] demonstrated the high predictive value of PITX2 DNA methylation for treatment with anthracyclines for triple-negative breast cancer patients, applying a qPCR-based research test. However, in this study, hypomethylation of PITX2 was associated with poor outcome, instead of hypermethylation as demonstrated in the present study and by others [10,11,13]. Triple-negative breast cancer is a heterogeneous disease, which may explain the opposite relationship of PITX2 DNA methylation. Subtypes on a molecular level have been proposed to further characterize triple-negative breast cancer, and despite some differences in the number of subtypes or classifying methods, the clinical relevance of the triple-negative breast cancer subtypes is, however, not yet defined. To explain the opposite relationship of PITX2 DNA methylation in triple-negative and ER-positive breast cancer, Aubele et al. [12] developed and presented a molecular-pathological hypothesis.
PITX2 DNA methylation not only predicts outcome in high-risk breast cancer patients treated with anthracycline-based chemotherapy, but also predicts outcome in tamoxifen-treated lymph node-negative breast cancer patients [11,13]. Furthermore, PITX2 DNA hypermethylation identifies breast cancer patients who have not received any systemic therapy as being at high risk for disease recurrence, hinting to the assumption that PITX2 methylation has prognostic value [28]. So far, there are no data available that demonstrate that PITX2 DNA methylation can predict outcome after taxanes or aromatase inhibitors. The studies available to date suggest that PITX2 DNA methylation has both prognostic and predictive value. In order to demonstrate that PITX2 DNA methylation is predictive of anthracycline efficacy, additional studies are required to investigate the effect in breast cancer patients treated with non-anthracycline regimens according to current treatment guidelines. The proposed study should utilize tissue specimens preferably from a randomized trial comparing both treatment modalities and, by applying a formal statistical test of the treatment-by-biomarker interaction, should be significant as described by Ballmann [29].
Standard of care according to guidelines is anthracycline-based chemotherapy. However, it is noteworthy that anthracycline-free regimens seem as effective. Shulman et al. [3] demonstrated (Cancer and Leukemia Group B (CALGB) 40101 trial) that a single-agent taxane is not inferior compared to doxorubicin plus cyclophosphamide in ER-positive, HER2-negative disease. In addition, in the Oncology Research Trial 9735, Jones et al. [30] even showed that docetaxel with cyclophosphamide is superior to doxorubicin and cyclophosphamide.
The present study represents a multicenter study using routinely collected and archived primary tumor tissue samples obtained from early-stage high-risk breast cancer patients who have been treated according to standard-of-care guidelines. Applying the revised Levels of Evidence (LoE) rules for categorization of biomarker studies published by Simon et al. [31], our applied test and the associated studies fulfill the criteria for LoE II. The present study does not meet the criteria for LoE Ib because our tumor samples were not part of a prospectively planned and conducted clinical trial. However, future studies including a prospective clinical trial in the neoadjuvant setting will be conducted to increase the LoE for PITX2 DNA methylation as a clinically useful predictive marker to support the treating physician concerning the selection of patients significantly benefiting from adjuvant anthracycline-based chemotherapy.
Among the high-risk breast cancer patients who have been treated with endocrine therapy in addition to anthracycline-based chemotherapy, PITX2 DNA methylation identifies a low-risk patient population (more than 60% of patients in the present study) that has a 5-year DFS probability of approximately 88%. Clinical trials have demonstrated that lymph node-positive, ER-positive and HER2-negative breast cancer patients treated with anthracyclines in combination with docetaxel or paclitaxel or paclitaxel in combination with gemcitabine showed an approximately 5-year DFS probability of 84% [32]. Therefore, further studies are warranted to support the notion that PITX2 DNA methylation might select patients who are sufficiently treated with anthracycline-based chemotherapy in combination with endocrine therapy, and for whom administration of taxanes can be omitted. This would spare patients significant side effects of a chemotherapy component that is unnecessary if any benefit is highly unlikely.
In conclusion, applying a robust, reproducible assay, we can now determine the methylation status of the PITX2 gene in gDNA extracted from FFPE primary breast cancer tissue specimens with excellent analytical validity. Importantly, patients with a low PITX2 DNA methylation status are sufficiently treated with anthracycline-based chemotherapy with or without additional endocrine therapy, proven by an extended DFS interval. In contrast, patients with high PITX2 DNA methylation status have a worse outcome while on anthracycline-based chemotherapy and should be considered for alternative chemotherapy regimens or the addition of taxanes to the regimen [22,23].
Disclosure Statement
O.G.W., G.S., R.N., M.A., and M.S. are employees of Therawis Diagnostics. J.L. and J.P. are employees of QIAGEN. J.W.M.M. receives funding from Sanofi and Philips Corp. O.W., R.N., M.S., and M.K. are equity owners of Therawis Diagnostics GmbH. J.L. and J.P. are stock owners of QIAGEN GmbH. C.T. is a member of the following advisory boards and receives financial remunerations: Amgen, Astra-Zeneca, Celgene, Daiichi Sankyo, Eisai, Lilly, MSD, Nanostring, Novartis, Pfizer, Puma, and Roche, and travel support from: Astra-Zeneca, Pfizer, Roche, and research support (including for clinical studies) from: Affymetrix, American Diagnostica, Amgen, Astra-Zeneca, Celgene, German Breast Group, Nanostring, Novartis, Pfizer, Roche. W.W. is a member of the following advisory boards: AstraZeneca, Novartis, BMS, MSD, Roche, Pfizer, and Takeda. The work was in part performed under a research agreement supported by Therawis Diagnostics.
Acknowledgements
This work has been partly supported by Therawis Diagnostics GmbH and a grant from the German Cancer Aid (DKH) to W.W. for the Transluminal B project in the framework of the DKH Translational Oncology Program. J.W.M.M. received support from Cancer Genomics Netherlands (CGC.nl), a consortium funded by the Netherlands Organization for Scientific Research (NWO). The funding sources had no impact or role in the study design, data collection, data analysis, interpretation and writing of the report.
References
- 1.Peto R, Davies C, Godwin J, Gray R, Pan HC, Clarke M, Cutter D, Darby S, McGale P, Taylor C, Wang YC, Bergh J, Di Leo A, Albain K, Swain S, Piccart M, Pritchard K. Comparisons between different polychemotherapy regimens for early breast cancer: meta-analyses of long-term outcome among 100,000 women in 123 randomised trials. Lancet. 2012;379:432–444. doi: 10.1016/S0140-6736(11)61625-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Turner N, Biganzoli L, Di Leo A. Continued value of adjuvant anthracyclines as treatment for early breast cancer. Lancet Oncol. 2015;16:e362–e369. doi: 10.1016/S1470-2045(15)00079-0. [DOI] [PubMed] [Google Scholar]
- 3.Shulman LN, Berry DA, Cirrincione CT, Becker HP, Perez EA, O'Regan R, Martino S, Shapiro CL, Schneider CJ, Kimmick G, Burstein HJ, Norton L, Muss H, Hudis CA, Winer EP. Comparison of doxorubicin and cyclophosphamide versus single-agent paclitaxel as adjuvant therapy for breast cancer in women with 0 to 3 positive axillary nodes: CALGB 40101 (Alliance) J Clin Oncol. 2014;32:2311–2317. doi: 10.1200/JCO.2013.53.7142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Basu M, Roy SS. Wnt/β-catenin pathway is regulated by PITX2 homeodomain protein and thus contributes to the proliferation of human ovarian adenocarcinoma cell, SKOV-3. J Biol Chem. 2013;288:4355–4367. doi: 10.1074/jbc.M112.409102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baylin SB. DNA methylation and gene silencing in cancer. Nat Clin Pract Oncol. 2005;2((suppl 1)):S4–S11. doi: 10.1038/ncponc0354. [DOI] [PubMed] [Google Scholar]
- 6.Franco D, Christoffels VM, Campione M. Homeobox transcription factor Pitx2: the rise of an asymmetry gene in cardiogenesis and arrhythmogenesis. Trends Cardiovasc Med. 2014;24:23–31. doi: 10.1016/j.tcm.2013.06.001. [DOI] [PubMed] [Google Scholar]
- 7.Lamba P, Hjalt TA, Bernard DJ. Novel forms of Paired-like homeodomain transcription factor 2 (PITX2): generation by alternative translation initiation and mRNA splicing. BMC Mol Biol. 2008;9:31. doi: 10.1186/1471-2199-9-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pillai SG, Dasgupta N, Siddappa CM, Watson MA, Fleming T, Trinkaus K, Aft R. Paired-like homeodomain transcription factor 2 expression by breast cancer bone marrow disseminated tumor cells is associated with early recurrent disease development. Breast Cancer Res Treat. 2015;153:507–517. doi: 10.1007/s10549-015-3576-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rowlands TM, Pechenkina IV, Hatsell SJ, Pestell RG, Cowin P. Dissecting the roles of beta-catenin and cyclin D1 during mammary development and neoplasia. Proc Natl Acad Sci U S A. 2003;100:11400–11405. doi: 10.1073/pnas.1534601100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hartmann O, Spyratos F, Harbeck N, Dietrich D, Fassbender A, Schmitt M, Eppenberger-Castori S, Vuaroqueaux V, Lerebours F, Welzel K, Maier S, Plum A, Niemann S, Foekens JA, Lesche R, Martens JWM. DNA methylation markers predict outcome in node-positive, estrogen receptor-positive breast cancer with adjuvant anthracycline-based chemotherapy. Clin Cancer Res. 2009;15:315–323. doi: 10.1158/1078-0432.CCR-08-0166. [DOI] [PubMed] [Google Scholar]
- 11.Maier S, Nimmrich I, Koenig T, Eppenberger-Castori S, Bohlmann I, Paradiso A, Spyratos F, Thomssen C, Mueller V, Nahrig J, Schittulli F, Kates R, Lesche R, Schwope I, Kluth A, Marx A, Martens JWM, Foekens JA, Schmitt M, Harbeck N, European Organisation for Research and Treatment of Cancer (EORTC) PathoBiology group DNA-methylation of the homeodomain transcription factor PITX2 reliably predicts risk of distant disease recurrence in tamoxifen-treated, node-negative breast cancer patients - technical and clinical validation in a multi-centre setting in collaboration with the European Organisation for Research and Treatment of Cancer (EORTC) PathoBiology group. Eur J Cancer. 2007;43:1679–1686. doi: 10.1016/j.ejca.2007.04.025. [DOI] [PubMed] [Google Scholar]
- 12.Aubele M, Schmitt M, Napieralski R, Paepke S, Ettl J, Absmaier M, Magdolen V, Martens JWM, Foekens JA, Wilhelm OG, Kiechle M. The predictive value of PITX2 DNA methylation for high-risk breast cancer therapy: current guidelines, medical needs, and challenges. Dis Markers. 2017;2017:4934608. doi: 10.1155/2017/4934608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harbeck N, Nimmrich I, Hartmann A, Ross JS, Cufer T, Grutzmann R, Kristiansen G, Paradiso A, Hartmann O, Margossian A, Martens JWM, Schwope I, Lukas A, Muller V, Milde-Langosch K, Nahrig J, Foekens J, Maier S, Schmitt M, Lesche R. Multicenter study using paraffin-embedded tumor tissue testing PITX2 DNA methylation as a marker for outcome prediction in tamoxifen-treated, node-negative breast cancer patients. J Clin Oncol. 2008;26:5036–5042. doi: 10.1200/JCO.2007.14.1697. [DOI] [PubMed] [Google Scholar]
- 14.Absmaier M, Napieralski R, Schuster T, Aubele M, Walch A, Magdolen V, Dorn J, Gross E, Harbeck N, Noske A, Kiechle M, Schmitt M. PITX2 DNA-methylation predicts response to anthracycline-based adjuvant chemotherapy in triple-negative breast cancer patients. Int J Oncol. 2018;52:755–767. doi: 10.3892/ijo.2018.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hayes DF. Considerations for implementation of cancer molecular diagnostics into clinical care. Am Soc Clin Oncol Educ Book. 2016;35:292–296. doi: 10.1200/EDBK_160236. [DOI] [PubMed] [Google Scholar]
- 16.Napieralski R, Schricker G, Piednoir E, Manner O, Bona A, Segalas S, Schueren E, Lauber J, Perkins J, Noske A, Magdolen V, Weichert W, Kiechle M, Wilhelm OG, Schmitt M. therascreen PITX2 RGQ PCR assay for the assessment of PITX2 DNA-methylation status to investigate the role of the transcription factor PITX2 and the regulation of the Wnt/β-catenin pathway in pathophysiological processes. Protoc Exch. 2018 DOI: 10.1038/protex.2018.022. [Google Scholar]
- 17.Hothorn T. Maxstat: maximally selected rank statistics; in R package version 0.7–14. The R Foundation for Statistical Computing. 2017 [Google Scholar]
- 18.Fine JP, Gray RJ. A proportional hazards model for the subdistribution of a competing risk. J Am Stat Assoc. 1999;94:496–509. [Google Scholar]
- 19.Cox DR, Oakes D. London: Chapman and Hall; 1984. Analysis of Survival Data. [Google Scholar]
- 20.Harrell FE. New York: Springer; 2001. Regression Modeling Strategies: With Applications to Linear Models, Logistic Regression, and Survival Analysis. [Google Scholar]
- 21.Schoenfeld D. Chi-squared goodness-of-fit tests for the proportional hazards regression model. Biometrika. 1980;67:145–153. [Google Scholar]
- 22.Goldhirsch A, Wood WC, Coates AS, Gelber RD, Thurlimann B, Senn HJ. Strategies for subtypes - dealing with the diversity of breast cancer: highlights of the St. Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2011. Ann Oncol. 2011;22:1736–1747. doi: 10.1093/annonc/mdr304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.AGO: Guidelines of the AGO Breast Committee www.ago-online.de.
- 24.Di Leo A, Desmedt C, Bartlett JM, Piette F, Ejlertsen B, Pritchard KI, Larsimont D, Poole C, Isola J, Earl H, Mouridsen H, O'Malley FP, Cardoso F, Tanner M, Munro A, Twelves CJ, Sotiriou C, Shepherd L, Cameron D, Piccart MJ, Buyse M. HER2 and TOP2A as predictive markers for anthracycline-containing chemotherapy regimens as adjuvant treatment of breast cancer: a meta-analysis of individual patient data. Lancet Oncol. 2011;12:1134–1142. doi: 10.1016/S1470-2045(11)70231-5. [DOI] [PubMed] [Google Scholar]
- 25.Desmedt C, Di Leo A, de Azambuja E. Multifactorial approach to predicting resistance to anthracyclines. J Clin Oncol. 2011;29:1578–1586. doi: 10.1200/JCO.2010.31.2231. [DOI] [PubMed] [Google Scholar]
- 26.Gupta N, Schmitt F, Grebhardt S, Mayer D. β-Catenin is a positive regulator of estrogen receptor-α function in breast cancer cells. Cancers (Basel) 2011;3:2990–3001. doi: 10.3390/cancers3032990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Imai Y, Ishikawa E, Asada S, Sugimoto Y. Estrogen-mediated post transcriptional down-regulation of breast cancer resistance protein/ABCG2. Cancer Res. 2005;65:596–604. [PubMed] [Google Scholar]
- 28.Nimmrich I, Sieuwerts AM, Meijer-van Gelder ME, Schwope I, Bolt-de Vries J, Harbeck N, Koenig T, Hartmann O, Kluth A, Dietrich D, Magdolen V, Portengen H, Look MP, Klijn JG, Lesche R, Schmitt M, Maier S, Foekens JA, Martens JWM. DNA hypermethylation of PITX2 is a marker of poor prognosis in untreated lymph node-negative hormone receptor-positive breast cancer patients. Breast Cancer Res Treat. 2008;111:429–437. doi: 10.1007/s10549-007-9800-8. [DOI] [PubMed] [Google Scholar]
- 29.Ballman KV. Biomarker: predictive or prognostic? J Clin Oncol. 2015;33:3968–3971. doi: 10.1200/JCO.2015.63.3651. [DOI] [PubMed] [Google Scholar]
- 30.Jones S, Holmes FA, O'Shaughnessy J, Blum JL, Vukelja SJ, McIntyre KJ, Pippen JE, Bordelon JH, Kirby RL, Sandbach J, Hyman WJ, Richards DA, Mennel RG, Boehm KA, Meyer WG, Asmar L, Mackey D, Riedel S, Muss H, Savin MA. Docetaxel with cyclophosphamide is associated with an overall survival benefit compared with doxorubicin and cyclophosphamide: 7-year follow-up of US Oncology Research Trial 9735. J Clin Oncol. 2009;27:1177–1183. doi: 10.1200/JCO.2008.18.4028. [DOI] [PubMed] [Google Scholar]
- 31.Simon RM, Paik S, Hayes DF. Use of archived specimens in evaluation of prognostic and predictive biomarkers. J Natl Cancer Inst. 2009;101:1446–1452. doi: 10.1093/jnci/djp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Swain SM, Tang G, Geyer CE, Jr, Rastogi P, Atkins JN, Donnellan PP, Fehrenbacher L, Azar CA, Robidoux A, Polikoff JA, Brufsky AM, Biggs DD, Levine EA, Zapas JL, Provencher L, Northfelt DW, Paik S, Costantino JP, Mamounas EP, Wolmark N. Definitive results of a phase III adjuvant trial comparing three chemotherapy regimens in women with operable, node-positive breast cancer: the NSABP B-38 trial. J Clin Oncol. 2013;31:3197–3204. doi: 10.1200/JCO.2012.48.1275. [DOI] [PMC free article] [PubMed] [Google Scholar]