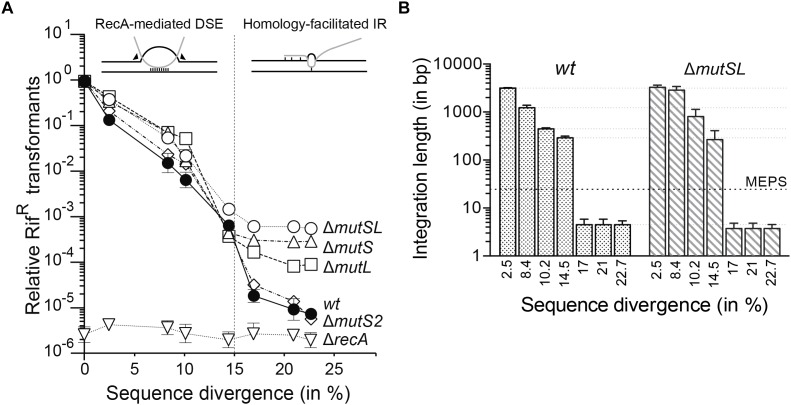
FIGURE 1.
Average chromosomal transformation frequencies as a function of sequence divergence from recipient. (A) Top insets, black lines represent recipient chromosomal DNA and the gray line, donor linear ssDNA. The vertical dashed line denotes MEPS. Left inset, when MEPS is found, the transformation heteroduplex forms a double-ended D-loop structure. Arrowheads indicate simultaneous incision by a putative D-loop resolvase to remove the recipient strand. Right inset, when MEPS is not found, RecA anchored at the recipient dsDNA facilitates spontaneous formation of short D-loops. Plasmid-borne rpoB482 DNA (0.1 μg DNA/ml) from different Bacillus species, with the selectable RifR mutation located at position 1443, was used to transform BG1359 (wt,  ) and its isogenic derivatives BG1393 (ΔmutSL,
) and its isogenic derivatives BG1393 (ΔmutSL,  ), BG1531 (ΔmutL,
), BG1531 (ΔmutL,  ), BG1393 (ΔmutSL plus pCB1018-borne mutL, Δ), BG1481 (ΔmutS2,
), BG1393 (ΔmutSL plus pCB1018-borne mutL, Δ), BG1481 (ΔmutS2,  ), and BG1633 (ΔrecA, ∇) competent cells. Sequence divergence values for the various Bacillus species were 0.04, 2.47, 8.35, 10.12, 14.52, 17.00, 20.83, and 22.74%. All data points are mean ± standard error of the mean (SEM) from three to five independent experiments. (B) Length of integration of donor DNA. The length of integrated DNA segments was determined by nucleotide sequence of the 2,997-bp rpoB482 DNA. Integration endpoints were defined by appearance of the predicted sequence of donor DNA and absence of expected mismatches in the recipient (integration midpoint). Distribution of integration endpoints up- and downstream of the RifR marker are shown for wt and ΔmutSL strains. The horizontal dashed black line denotes the MEPS and dotted gray lines denotes the small differences.
), and BG1633 (ΔrecA, ∇) competent cells. Sequence divergence values for the various Bacillus species were 0.04, 2.47, 8.35, 10.12, 14.52, 17.00, 20.83, and 22.74%. All data points are mean ± standard error of the mean (SEM) from three to five independent experiments. (B) Length of integration of donor DNA. The length of integrated DNA segments was determined by nucleotide sequence of the 2,997-bp rpoB482 DNA. Integration endpoints were defined by appearance of the predicted sequence of donor DNA and absence of expected mismatches in the recipient (integration midpoint). Distribution of integration endpoints up- and downstream of the RifR marker are shown for wt and ΔmutSL strains. The horizontal dashed black line denotes the MEPS and dotted gray lines denotes the small differences.