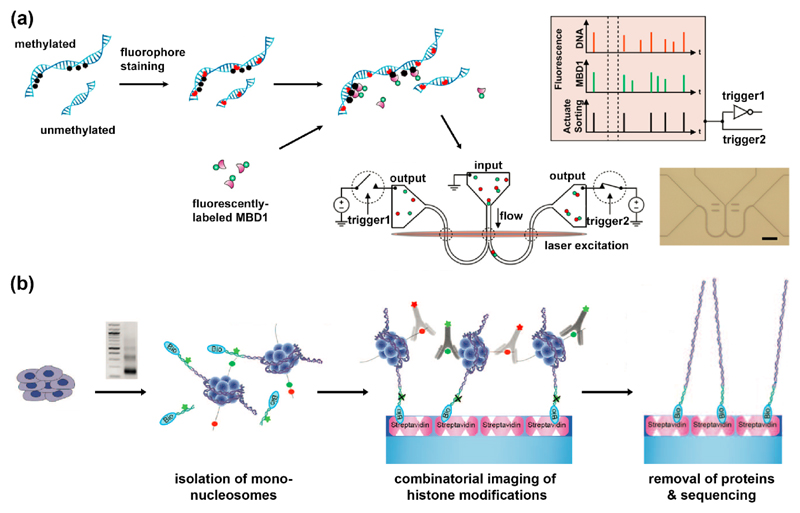
Figure 2.
Affinity-based methods for optical detection of epigenetic marks. (a) Single-molecule detection of DNA methylation sites. Methylation sites on stained DNA are labeled with fluorescent MBD1 protein. Two-color labeled DNA is then loaded on a nano-fluidic device, detected and sorted according to its fluorescent signature [15]. (b) Single-molecule detection of post translational modifications on nucleosomes. Nucleosomes are prepared by MNase digestion. DNA ends are ligated to fluorescent biotinylated adaptors, purified and captured on PEG-streptavidin-coated slides. Attached nucleosomes are incubated with fluorescently-labeled antibodies to histone modifications which can be then imaged. Finally, histones are removed, and their genomic position is determined by single-molecule sequencing-by-synthesis [18]. Reprinted with permission from AAAS.