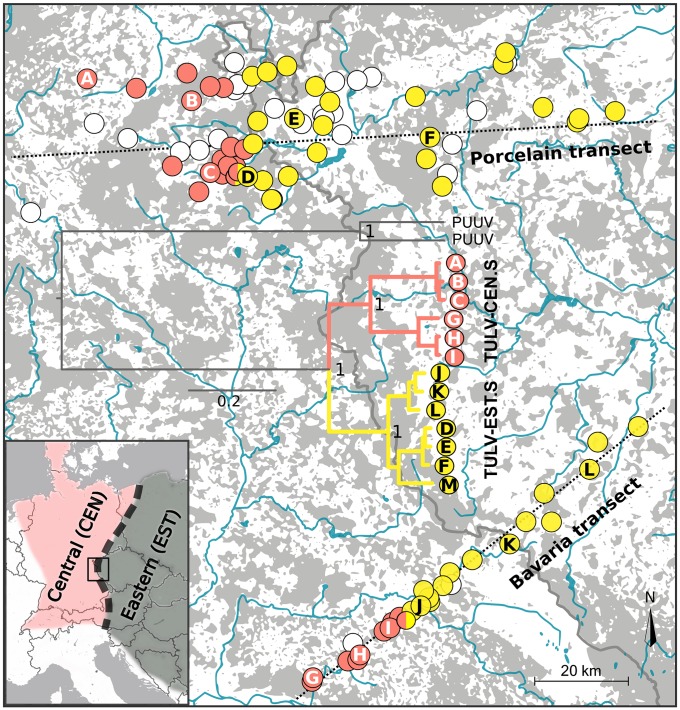
Fig 1. Distribution of two highly diverged TULV clades in a hybrid zone of their rodent reservoir host, the common vole M. arvalis.
TULV-CEN.S (red) and TULV-EST.S (yellow) virus clades only co-occurred in two localities (bicolored) in the Porcelain transect and one in the Bavaria transect. The insert shows the position of the study region in Central Europe and the approximate distribution of the Central and Eastern evolutionary lineages in the common vole. Phylogenetic relationships based on complete TULV genome sequences are shown for samples from 12 localities across the host hybrid zone (A–L). The only published genome sequence from Eastern Czech Republic (M) was included for reference (see map in Fig 2). PUUV was used as outgroup (see S7 Table for accession numbers), and posterior probabilities of major nodes are indicated. Host sampling localities in which no TULV infections were detected are shown as white circles. Gray areas on the map represent forest (unsuitable habitat for common voles), and major water bodies are shown in blue. The border between Germany and the Czech Republic is indicated with a gray line. CEN, Central host lineage; EST, Eastern host lineage; PUUV, Puumala orthohantavirus; TULV, Tula orthohantavirus; TULV-CEN.S, Central South TULV; TULV-EST.S, Eastern South TULV.