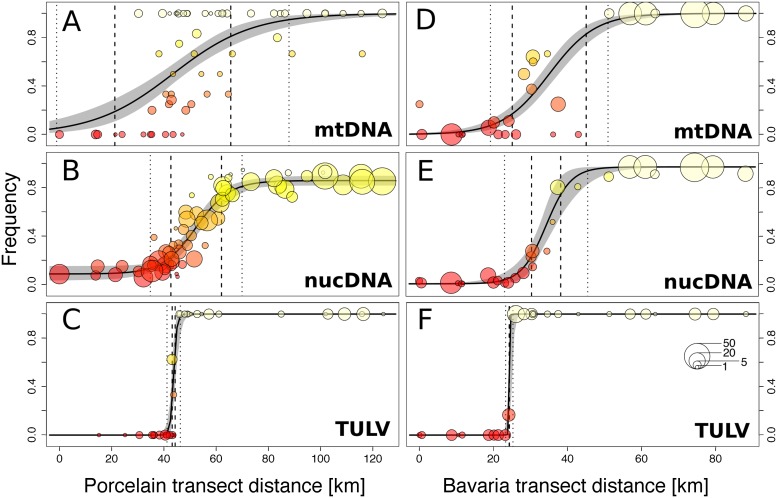
Fig 3. Geographic clines representing the transition between evolutionary lineages in the rodent host and phylogenetic clades in TULV along the Porcelain (A, B, and C) and Bavaria (D, E, and F) transects.
The contact between the Central and the Eastern common vole lineages was characterized based on mtDNA (A and D) and nucDNA (B and E) markers, whereas for TULV clades partial S-segment sequences were phylogenetically assigned to TULV-CEN.S and TULV-EST.S clades (C and F; see Fig 2). The frequency per locality of Eastern host lineage (A, B, D, and E) and TULV-EST.S virus clade (C and F) is displayed starting from the western-most locality. Symbol sizes correspond to the number of samples and symbol colors to the genotype frequency per location. 95% credible cline regions are shown in gray. Cline widths are shown around the estimated cline center, with dotted and dashed lines indicating upper and lower width estimates (two log-likelihood support limits), respectively. mtDNA, mitochondrial DNA; nucDNA, nuclear DNA; TULV, Tula orthohantavirus; TULV-CEN.S, Central South TULV; TULV-EST.S, Eastern South TULV.