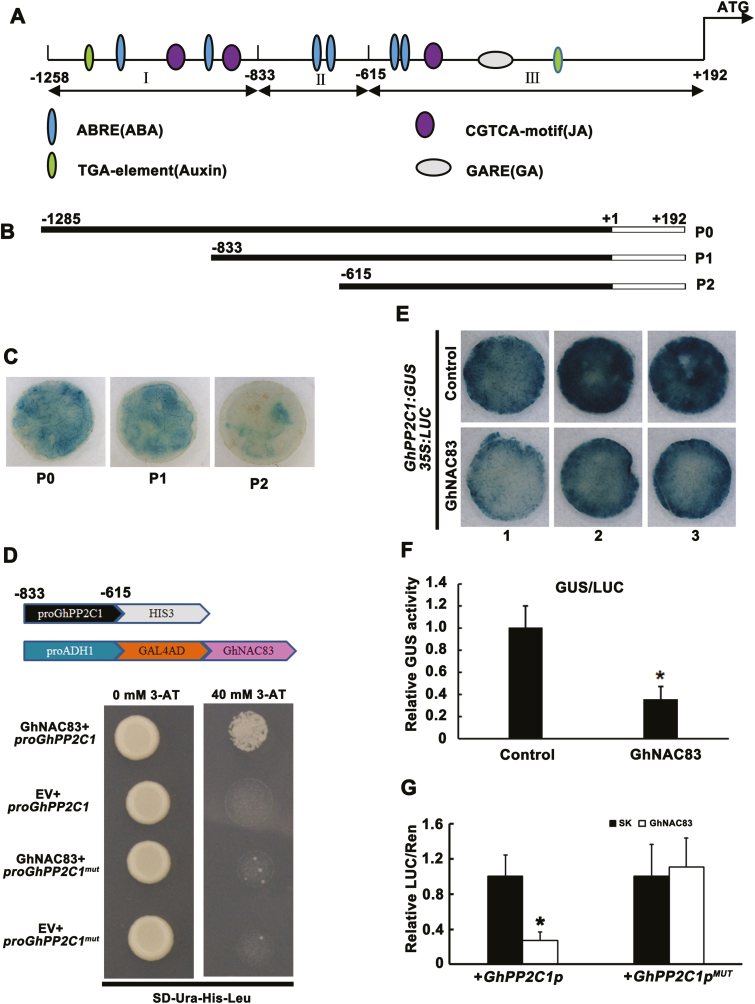
Fig. 5.
GhNAC83 binds the GhPP2C1 promoter and inhibits its transcription. (A) The distribution of cis-elements in the GhPP2C1 promoter. (B) Truncations of the GhPP2C1 promoter used in transient assays. (C) Deletion of base pairs –833 to –615 in the GhPP2C1 promoter (P2 construct shown in B) dramatically affects its activity in transient N. benthamiana assays. Three biological replicates were conducted and showed similar results. One biological replicate of leaf discs is shown. (D) The interaction of GhNAC83 and the GhPP2C1 promoter in yeast one-hybrid assays. The empty prey vector (pDEST-GAD424) was used as the control. The mutagenesis of NAC-binding sites (Supplementary Fig. S3) in the GhPP2C1 promoter (proGhPP2C1mut) was also tested. The interaction between the GhNAC83 protein and the GhPP2C1 promoter was determined by cell growth on synthetic dropout nutrient medium lacking Ura, His, and Leu, and containing 40mM 3-AT. (E) GhNAC83 represses GhPP2C1 promoter activity in transient expression assays in N. benthamiana leaves. 35S:GhNAC83 was used as an effector and GhPP2C1:GUS was used as a reporter. 35S:LUC was used as an internal control. GUS stains were performed on the third day post-infiltration. (F) The relative GUS activity (GUS/LUC) indicates that GhNAC83 represses GhPP2C1 transcription in planta. Three biological replicates were performed and are shown with the SD. (G) The interaction of GhNAC83 with the GhPP2C1 promoter using a dual-luciferase reporter assay in N. benthamiana leaves. A fragment of GhPP2C1’s promoter (base pairs +192 to –1285) was used in this assay. Mutated sites in GhPP2C1pMUT are shown in Supplementary Fig. S3. The empty vector (pGreenII 62-SK) was used as a control. Data are shown as the average of three biological replicates with the SD (n=5 leaves), *P<0.05. (This figure is available in color at JXB online.)