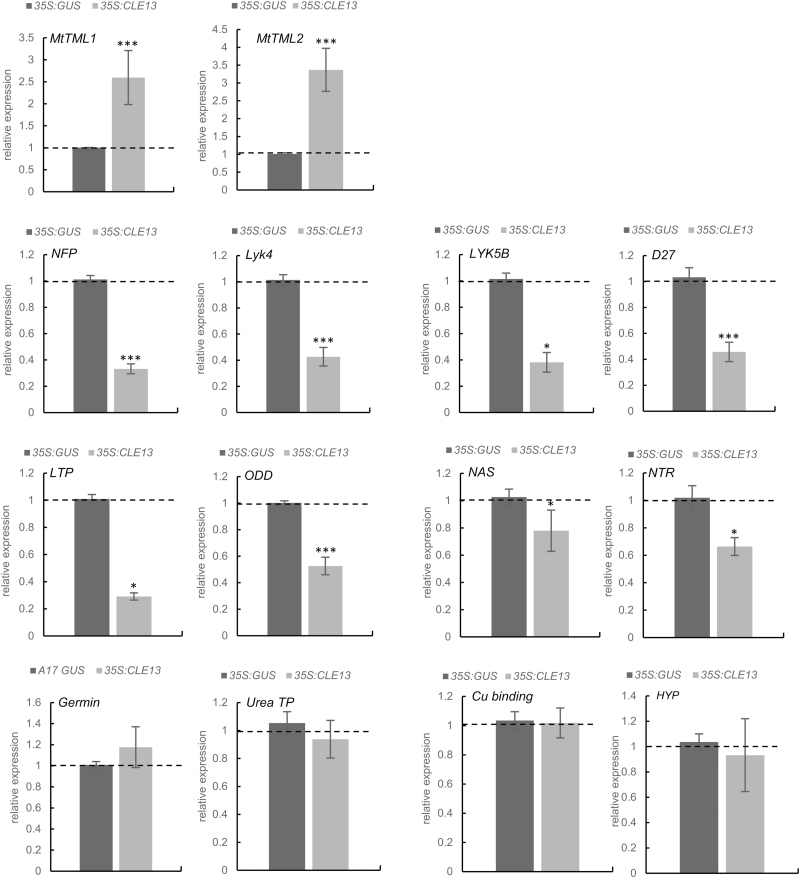
Fig. 1.
qRT–PCR validation of genes differentially expressed in 35S:MtCLE13 roots. Expression analysis by qRT–PCR identified different genes as deregulated in 35S:MtCLE13 roots compared with 35S:GUS (control) roots with the microarray approach (see Suppementary Dataset S1). Gene expression was calibrated relative to the 35S:GUS control roots to highlight fold changes, as indicated by the dotted line. Error bars represent the SE of the mean of at least three biological repeats (n>15 plants per biological replicate). *P<0.05; **P<0.01; ***P<0.001 indicate significant differences between the two genotypes as measured with an ANOVA mixed model with a Tukey’s post-hoc comparison. The gene expression levels were calibrated against the expression found in the 35S:GUS control roots. TML1 and TML2, Too Much Love 1 and 2; NFP, Nod Factor perception; LYK4, LysM receptor-like kinase (RLK) 4; LYK5B, LysM-RLK 5B; D27, β-carotene isomerase; LTP, lipid transfer protein; ODD, oxoglutarate-dependent dioxygenase; NAS, nicotianamide synthase; NTR, MtNRT2.3 high-affinity nitrate transporter; Urea TP, urea transporter; Cu-binding, copper-binding protein; HyP, hypothetical protein.