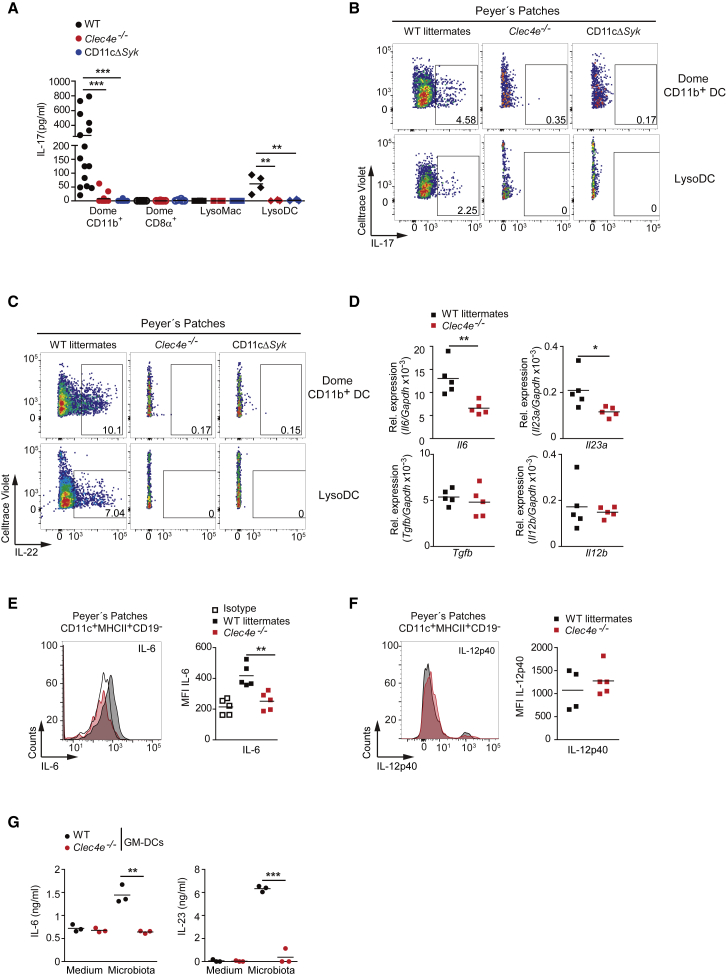
Figure 3.
DCs from PPs Instruct Mincle- and Syk-Dependent Th17 Differentiation
(A–C) Naive OT-II T cells were co-cultured for 3 days with dome CD11b+ DCs, CD8α+ DCs, LysoMacs, or LysoDCs from the indicated genotypes, sorted from PPs (1:1 ratio), and loaded with OVA323–339 peptide. (A) IL-17 secretion by ELISA. Each dot represents an independent co-culture where myeloid cells were from different mice from two independent experiments. (B and C) Representative FACS plots of IL-17 (B) and IL-22 (C) intracellular staining after OTII re-stimulation.
(D) Il6, Il23a, Tgfb, and Il12b transcripts in PPs of the indicated genotypes by qPCR; they were normalized to Gapdh.
(E and F) Analysis of IL-6 (E) and IL-12p40 (F) intracellular staining in CD11c+MHC-II+CD19− cells from PPs in the indicated genotypes. Shown on the left are representative histograms. On the right is MFI of staining.
(G) ELISA of IL-6 and IL-23 production by sorted GM-DCs from WT and Mincle-deficient (Clec4e−/−) mice untreated (medium) or stimulated with gut microbiota (10:1 DC ratio) for 12 h.
Data represent two independent pooled experiments (A) or one representative experiment of at least two performed (B–G). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 (A: one-way ANOVA and Bonferroni post hoc test; D, E, and G: unpaired two-tailed Student’s t test). See also Figure S3.