Abstract
Background
The present study carried out a meta‐analysis to investigate whether the interleukin‐1 receptor antagonist (IL‐1RN) VNTR polymorphism and three IL‐10 single‐nucleotide polymorphisms (SNPs) rs1800896, rs3021097, and rs1800872 are associated with psoriasis risk.
Methods
Wanfang, China National Knowledge Infrastructure, Medline, and PubMed databases were searched for potential studies published until 2 November 2017. Forest plots were generated.
Results
Thirteen case–control studies were included in the review. The results of meta‐analyses revealed no association of the IL‐1RN*2 allele with psoriasis in the overall populations (odds ratio [OR] = 1.16, 95% confidence intervals [CI]: 0.89–1.50, p = 0.279), Asians (OR = 1.27, 95% CI: 0.73–2.23, p = 0.403), and Caucasians (OR = 1.04, 95% CI: 0.88–1.23, p = 0.669). Under the allelic model, there was no statistically significant association of psoriasis with the IL‐10 SNPs rs1800896 (G allele vs. A allele: OR = 1.03, 95% CI: 0.90–1.18, p = 0.639), rs3021097 (C allele vs. T allele: OR = 1.17, 95% CI: 0.88–1.56, p = 0.288), and rs1800872 (C allele vs. A allele: OR = 1.01, 95% CI: 0.81–1.25, p = 0.951). No publication bias was found by Egger's test and Begg's funnel plots.
Conclusion
Current published studies fail to support an association of the IL‐1RN VNTR polymorphism and IL‐10 SNPs rs1800896, rs3021097, and rs1800872 with psoriasis risk.
Keywords: interleukin‐1 receptor antagonist, interleukin‐10, polymorphism, psoriasis
1. INTRODUCTION
Psoriasis is a common inflammatory disease of the skin affecting 2% of the population (Boehncke & Schon, 2015). Plaque psoriasis, the most common variant of psoriasis, is characterized by inflamed, red skin covered by a silvery white scale (Boehncke & Schon, 2015). Psoriasis is an independent risk factor for mortality and contributes to numerous comorbid conditions, including rheumatological arthritis, depression, cardiovascular disease, and diabetes (Kim, Jerome, & Yeung, 2017). At present, there is no cure for psoriasis and there are no specific markers that can accurately predict the development of psoriasis. Although psoriasis pathogenesis remains poorly understood, it is accepted by most dermatologists that psoriasis arises via the interactions of genetic, immunological, and environmental factors (Mahil, Capon, & Barker, 2016). Currently, more than 40 independent loci are associated with the susceptibility to psoriasis.
Interleukin‐1 receptor antagonist (IL‐1Ra) is a member of the IL‐1 family and an important anti‐inflammatory cytokine. IL‐1Ra competitively blocks the effects of IL‐1αand IL‐1β by binding to the IL‐1 receptor. An inbalance between IL‐1 and IL‐1Ra is associated with increased production of pro‐inflammatory cytokines and the development of inflammatory disorders (Mistry, Savic, & Hilst, 2017). IL‐1Ra‐deficient mice spontaneously developed a dermatitis that histologically resembled human psoriasis (Nakajima et al., 2010; Shepherd, Little, & Nicklin, 2004). In a human coculture model with keratinocytes and autologous T cells, application of a recombinant human form of IL‐1Ra significantly reduced pro‐inflammatory cytokine production (Renne, Schafer, Werfel, & Wittmann, 2010). In addition, the levels of pro‐inflammatory cytokines were significantly up‐regulated in lesional psoriatic epidermis due to decreased formation and secretion of IL‐1Ra (Debets et al., 1997; Kristensen et al., 1992). In intron 2 of the IL‐1Ra gene (IL‐1RN), there is an 86‐basepair variable number tandem repeat (VNTR) polymorphism. The IL‐1RN allele 2 is thought to result in higher IL‐1Ra release (Bid, Manchanda, & Mittal, 2006). Since IL‐1Ra is involved in the inflammatory responses within psoriatic plaques, dermatologists have carried out genetic studies to assess the relationship of the IL‐1RN VNTR polymorphism with psoriasis risk. A quantitative summary of their findings is needed.
IL‐10 is an anti‐inflammatory cytokine synthesized by monocytes, macrophages, and lymphocytes as a response to inflammation (Fontoura et al., 2015). It is able to inhibit antigen presentation and suppress the synthesis and function of a number of pro‐inflammatory cytokines, including tumor necrosis factor‐α (TNF‐α), IL‐6, and IL‐1 (Rutz & Ouyang, 2016). IL‐10 mRNA expression was dramatically decreased in the psoriatic lesions compared with the normal skin tissues (Cheng et al., 2001). In addition, levels of IL‐10‐producing regulatory B cells (B10 cells) were decreased in patients with psoriasis (Hayashi et al., 2017). Given that IL‐10 played a protective role in inflammation development, adoptive transfer of B10 cells effectively suppressed imiquimod‐induced skin inflammation in a mouse model of psoriasis (Yanaba et al., 2013). Moreover, administration of recombinant IL‐10 significantly reduced the severity of psoriasis in humans (Asadullah et al., 1999). Three single‐nucleotide polymorphisms (SNPs) at ‐592A/C (rs1800872), ‐819C/T (rs3021097), and ‐1082A/G (rs1800896) in the IL‐10 promoter region have recently been evaluated in a number of genetic association studies regarding their relationship with psoriasis susceptibility, but the findings of these studies are contradictory.
Both IL‐1Ra and IL‐10 are important anti‐inflammatory cytokines in psoriasis pathogenesis. Our study seeks to systematically review the literature and meta‐analyze the results of case–control studies on the relation of IL‐1RN and IL‐10 polymorphisms with psoriasis risk.
2. METHODS
2.1. Ethical compliance
Ethics approval was waived because this study does not involve any human participants or animals.
2.2. Systematic literature search
On 2nd of November 2017, two reviewers carried out independent electronic searches on the Wanfang, China National Knowledge Infrastructure (CNKI), Medline, and PubMed databases covering the entire period of each database. The following algorithm was applied: “(interleukin‐1 OR interleukin‐10 OR cytokine) AND (psoriasis OR genetics OR polymorphism).” Results were limited to studies in English or Chinese, featuring human participants and with abstracts available. We supplemented these searches by searching review articles and reference lists of the included studies. We did not contact study authors to request additional data. Disagreements were solved by discussion and consensus.
2.3. Inclusion/exclusion criteria
Studies were assessed against the following inclusion criteria: (a) assessing the relation of the IL‐1RN polymorphism or the IL‐10 polymorphisms (rs1800896, rs3021097, and rs1800872) with psoriasis risk; (b) used validated measures to determine the presence of psoriasis; (c) the study was case–control designed; (d) all data were original; (e) sufficient information on odds ratios (ORs) with their 95% confidence intervals (95% CIs). Exclusion criteria were as follows: (a) not case–control design; (b) not an original paper; (c) performed on animals or human cell lines; (d) studies based on only cases; (e) not offering essential data. Additionally, studies with potentially overlapping cohorts were excluded from the quantitative synthesis. In such cases, we included the study with the largest sample size.
2.4. Data extraction
Data extraction was conducted by two investigators (Ju Qiao and Qian‐Nan Jia). The senior author (Hong‐Zhong Jin) was involved in consulting for the eligibility of a study if a divergence between the two data‐extracting investigators existed. From the finally selected papers, data relating to study characteristics for the following variables were extracted: first author, journal name, publication year, location of study, ethnicity, male percentage, number of study subjects, the frequencies of genotypes or alleles in case and control groups, genotyping method, and Hardy–Weinberg equilibrium (HWE) status.
2.5. Statistical analyses
All statistical analyses were conducted using Stata version 10.0. To assess the relation of the IL‐1RN VNTR polymorphism and the IL‐10 variants with susceptibility to psoriasis, pooled ORs and their corresponding 95% CIs were assessed. Between‐study heterogeneity was evaluated using Cochrane’s Q statistic. There are two statistical methods for meta‐analysis: the fixed‐effects model and the random‐effects model. If the Q test was statistically significant (p < 0.10), estimates were pooled using random‐effects models (SanGiovanni, Berkey, Dwyer, & Colditz, 2000). When between‐study heterogeneity was not significant, the overall effects were calculated using the fixed‐effects model (Mantel & Haenszel, 1959). HWE in controls was calculated by chi‐square test. Funnel plots were drawn as a check for potential publication bias. We also used Egger's asymmetry test to evaluate publication bias. The significance level was set at p < 0.05, except for test of heterogeneity.
3. RESULTS
3.1. Study characteristics
The execution of the search strategy initially resulted in 129 studies after duplicates were removed. Subsequently, 109 studies were excluded at the title/abstract level, and 20 full‐text papers were checked for eligibility. After careful evaluation, a total of 13 studies were included in the review (Chang et al., 2007; Craven et al., 2001; Indhumathi et al., 2017; Karam, Zidan, & Khater, 2014; Li, Dong, Zhu, Yu, & Wang, 1999; Liu, Zhang, Yang, & Xu, 1998; Moorchung, Vasudevan, Chatterjee, Mani, & Grewal, 2015; Peddle, Butt, Snelgrove, & Rahman, 2005; Peng & Wang, 1999; Reich et al., 2002, 1999 ; Tarlow et al., 1997; Wongpiyabovorn et al., 2008). We did not find any additional studies by hand‐searching the reference lists of the included studies. Figure 1 summarizes the process of study selection. The qualified publications included in our meta‐analysis were published between 1997 and 2017. The sample sizes ranged from 60 to 720 participants. Tables 1 and 2 present the characteristics of the individual studies. Figure 2 shows sample sizes of reports over time. We did not find any studies violating HWE in controls.
Figure 1.
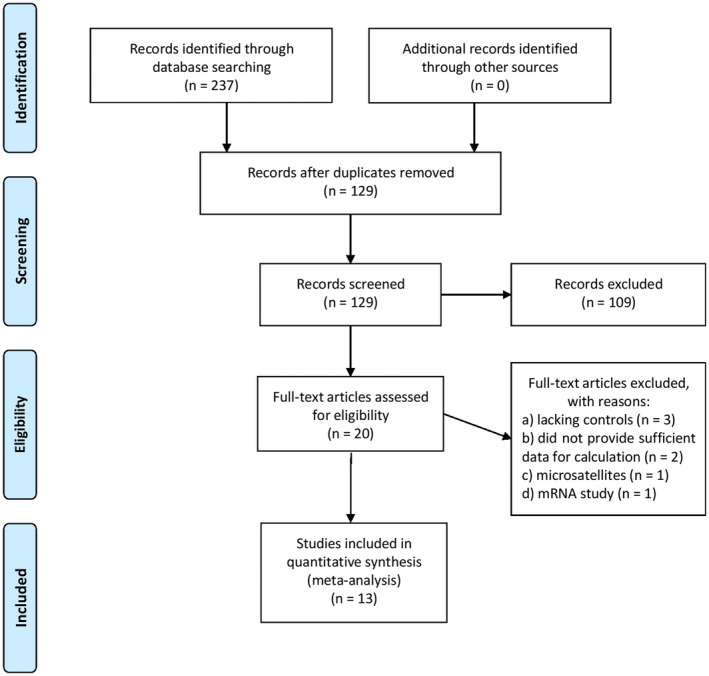
Process of study selection
Table 1.
Characteristics of the studies evaluating the relationship between the IL‐1RN VNTR polymorphism and psoriasis risk
| Author | Journal | Year | Country | Ethnicity | Controls | Cases | Male in controls (%) | Male in cases (%) | Genotyping method | Controls selection | HWE |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Tarlow | Br J Dermatol | 1997 | UK and Germany | Caucasians | 331 | 271 | Not available | Not available | PCR assay | Not available | Yes |
| Liu | Natl Med J China | 1998 | China | Asians | 130 | 82 | Not available | 56.1 | PCR assay | Hospital | Yes |
| Peng | Immunol J | 1999 | China | Asians | 85 | 38 | Not available | 63.2 | PCR assay | Population | Yes |
| Li | J Clin Dermatol | 1999 | China | Asians | 30 | 30 | Not available | 53.3 | PCR assay | Not available | Yes |
| Reich | J Invest Dermatol | 2002 | Germany | Caucasians | 345 | 231 | 52.8 | 65.4 | PCR assay | Hospital | Yes |
| Peddle | Ann Rheum Dis | 2005 | Canada | Caucasians | 95 | 226 | Not available | 52.2 | PCR assay | Not available | Yes |
| Chang | Br J Dermatol | 2007 | China | Asians | 210 | 272 | Not available | 64.7 | PCR assay | Hospital | Yes |
| Moorchung | Indian J Dermatol | 2015 | India | Asians | 243 | 112 | Not available | 56.3 | PCR assay | Hospital | Yes |
HWE: Hardy–Weinberg equilibrium; IL‐1RN VNTR: interleukin‐1 receptor antagonist variable number tandem repeat; PCR: polymerase chain reaction; UK: United Kingdom.
Table 2.
Characteristics of the included studies assessing the association between the IL‐10 polymorphisms and psoriasis risk
| Author | Journal | Year | Country | Ethnicity | Controls | Cases | Male in controls (%) | Male in cases (%) | Genotyping method | Selection of controls | IL‐10 polymorphism |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Reich | J Invest Dermatol | 1999 | Germany | Caucasian | 123 | 151 | 54.5 | 63.6 | Sequencing analysis | Population | rs1800896 |
| Craven | Br J Dermatol | 2001 | UK | Caucasian | 330 | 78 | Not available | 41.7 | PCR assay | Hospital | rs1800896 |
| Peddle | Ann Rheum Dis | 2005 | Canada | Caucasian | 95 | 226 | Not available | 52.2 | PCR assay | Not available | rs1800896 |
| Chang | Br J Dermatol | 2007 | China | Asian | 210 | 272 | Not available | 64.7 | Sequencing analysis | Hospital | rs1800896, rs3021097, rs1800872 |
| Wongpiyabovorn | Clin Exp Dermatol | 2008 | Thailand | Asian | 155 | 139 | 16.1 | 56.8 | PCR‐RFLP | Population | rs1800896 and rs1800872 |
| Karam | Cytokine | 2014 | Egypt | Caucasian | 120 | 110 | 35.8 | 24.5 | PCR‐RFLP | Not available | rs1800896 |
| Indhumathi | Hum Immunol | 2017 | India | Asian | 360 | 360 | 80.0 | 81.1 | TaqMan 5’ allele discrimination assay | Not available | rs1800896 and rs3021097 |
IL‐10: interleukin‐10; PCR: polymerase chain reaction; PCR‐RFLP: polymerase chain reaction–restriction fragment length polymorphism; UK: United Kingdom.
Figure 2.
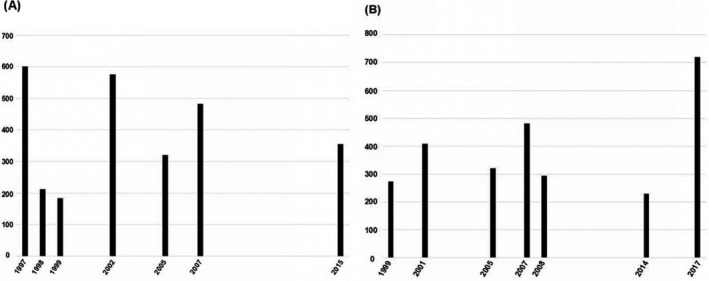
(a) Sample sizes of the studies assessing the interleukin‐1 receptor antagonist variable number tandem repeat polymorphism and risk of psoriasis from 1997 to 2015; (b) Sample sizes of the studies assessing the interleukin‐10 polymorphism rs1800896 from 1999 to 2017
3.2. No association of IL‐1RN VNTR polymorphism with psoriasis
Five studies evaluating the IL‐1RN VNTR polymorphism was performed in Asians (Chang et al., 2007; Li et al., 1999; Liu et al., 1998; Moorchung et al., 2015; Peng & Wang, 1999), and three in Caucasians (Peddle et al., 2005; Reich et al., 2002; Tarlow et al., 1997). Five studies provided genotype data for the polymorphism (Li et al., 1999; Liu et al., 1998; Moorchung et al., 2015; Peng & Wang, 1999; Reich et al., 2002), whereas three studies only contained information on allele frequency (Chang et al., 2007; Peddle et al., 2005; Tarlow et al., 1997). There was significant between‐study heterogeneity (Table 3). Random‐effect meta‐analyses combining the data did not identify any associations of the IL‐1RN VNTR polymorphism with psoriasis under four genetic models (dominant model: OR = 1.33, 95% CI: 0.76–1.35, p = 0.317; recessive model: OR = 1.77, 95% CI: 0.65–4.82, p = 0.262; additive model: OR = 1.80, 95% CI: 0.64–5.09, p = 0.266; allelic model: OR = 1.16, 95% CI: 0.89–1.50, p = 0.279; Table 3 and Figure 3). Then, subgroups analyses by different ethnicity (Asians and Caucasians) were carried out. The results suggested no significant association between the IL‐1RN VNTR polymorphism and psoriasis risk under any of the four genetic models in each ethnic group (Table 3 and Figure 3).
Table 3.
Summary of results for meta‐analyses
| Polymorphism | Dominant modela | Recessive modelb | Additive modelc | Allelic modeld | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | p value of the z test | p derived from heterogeneity test | OR (95% CI) | p value of the z test | p derived from heterogeneity test | OR (95% CI) | p value of the z test | p derived from heterogeneity test | OR (95% CI) | p value of the z test | p derived from heterogeneity test | |
| IL ‐1RN VNTR | ||||||||||||
| All (n = 8) | 1.33 (0.76–2.35) | 0.317 | 0.021 | 1.77 (0.65–4.82) | 0.262 | 0.001 | 1.80 (0.64–5.09) | 0.266 | 0.005 | 1.16 (0.89–1.50) | 0.279 | 0.003 |
| Asians (n = 5) | 1.53 (0.59–3.98) | 0.379 | 0.019 | 2.38 (0.63–9.02) | 0.201 | 0.003 | 2.65 (0.55–12.87) | 0.226 | 0.006 | 1.27 (0.73–2.23) | 0.403 | 0.001 |
| Caucasians (n = 3) | 0.98 (0.70–1.36) | 0.881 | Not applicable | 0.83 (0.45–1.55) | 0.566 | Not applicable | 0.84 (0.44–1.58) | 0.583 | Not applicable | 1.04 (0.88–1.23) | 0.669 | 0.677 |
| IL ‐10 rs1800896 | ||||||||||||
| All (n = 7) | 1.05 (0.85–1.29) | 0.672 | 0.184 | 1.18 (0.73–1.89) | 0.438 | 0.098 | 1.24 (0.76–2.04) | 0.395 | 0.147 | 1.03 (0.90–1.18) | 0.639 | 0.122 |
| Asians (n = 3) | 0.96 (0.73–1.27) | 0.771 | 0.278 | 1.13 (0.56–2.28) | 0.733 | 0.494 | 1.09 (0.54–2.21) | 0.815 | 0.467 | 0.93 (0.75–1.16) | 0.513 | 0.276 |
| Caucasians (n = 4) | 1.17 (0.85–1.61) | 0.331 | 0.123 | 1.19 (0.60–2.34) | 0.623 | 0.025 | 1.30 (0.62–2.71) | 0.492 | 0.046 | 1.10 (0.93–1.31) | 0.262 | 0.111 |
| IL ‐10 rs3021097 | ||||||||||||
| All (n = 2) | Not applicable | Not applicable | Not applicable | Not applicable | Not applicable | Not applicable | Not applicable | Not applicable | Not applicable | 1.17 (0.88–1.56) | 0.288 | 0.092 |
| IL ‐10 rs1800872 | ||||||||||||
| All (n = 2) | Not applicable | Not applicable | Not applicable | Not applicable | Not applicable | Not applicable | Not applicable | Not applicable | Not applicable | 1.01 (0.81–1.25) | 0.951 | 0.895 |
CI, confidence interval; IL‐1RN VNTR, interleukin‐1 receptor antagonist variable number tandem repeat; IL‐10, interleukin‐10; OR, odds ratio.
Dominant model: 22 + 2L versus LL for the IL‐1RN VNTR polymorphism; GG + GA versus AA for rs1800896.
Recessive model: 22 versus 2L + LL for the IL‐1RN VNTR polymorphism; GG versus GA + AA for rs1800896.
Additive model: 22 versus 2L versus LL for the IL‐1RN VNTR polymorphism; GG versus GA versus AA for rs1800896.
Allelic model: 2 allele versus L allele for the IL‐1RN VNTR polymorphism; G allele versus A allele for rs1800896; C allele versus T allele for rs3021097; C allele versus A allele for rs1800872.
Figure 3.
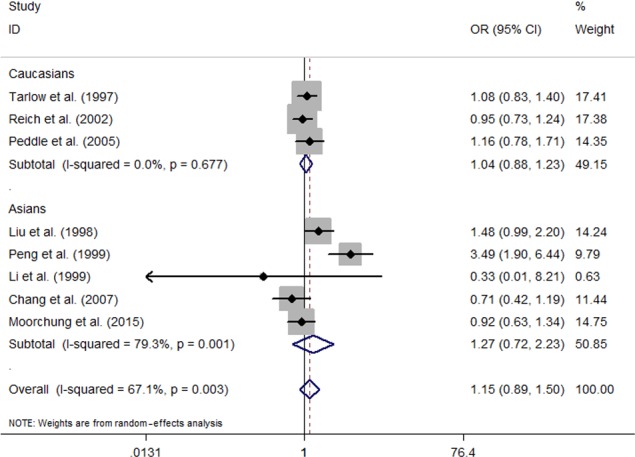
Forest plot of meta‐analysis with data on the association between the interleukin‐1 receptor antagonist variable number tandem repeat polymorphism and risk of psoriasis under allelic model
3.3. No association of IL‐10 polymorphisms with psoriasis
Seven studies provided summaries of data on the IL‐10 SNP rs1800896 (Chang et al., 2007; Craven et al., 2001; Indhumathi et al., 2017; Karam et al., 2014; Peddle et al., 2005; Reich et al., 1999; Wongpiyabovorn et al., 2008). No evidence for heterogeneity was observed across all studies (Table 3). Pooled results revealed no significant association of rs1800896 with psoriasis risk under any of the comparison models (dominant model: OR = 1.05, 95% CI: 0.85–1.29, p = 0.672; recessive model: OR = 1.18, 95% CI: 0.73–1.89, p = 0.438; additive model: OR = 1.24, 95% CI: 0.76–2.04, p = 0.395; allelic model: OR = 1.03, 95% CI: 0.90–1.18, p = 0.639; Table 3 and Figure 4). When studies were subgrouped by ethnicity, the relationship between rs1800896 and psoriasis was not significant in Asian or Caucasian populations (Table 3 and Figure 4).
Figure 4.
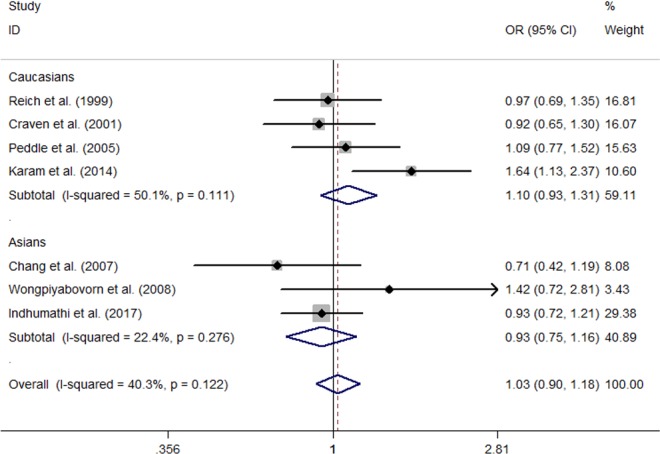
Forest plot of meta‐analysis with data on the association between the interleukin‐10 polymorphism rs1800896 under allelic model
Two studies investigated the relation between the IL‐10 SNP rs3021097 (Chang et al., 2007; Indhumathi et al., 2017) and psoriasis risk. Under allelic contrast, the summary OR was 1.17 (95% CI: 0.88–1.56, p = 0.288), suggesting no significant association with psoriasis (Table 3). Significant heterogeneity was identified (Table 3).
Two studies evaluated the association of the IL‐10 SNP rs1800872 with psoriasis risk using data on allelotype (Chang et al., 2007; Wongpiyabovorn et al., 2008). Meta‐analysis of fixed‐effects model provided an estimated OR of 1.01 (95% CI: 0.81–1.25, p = 0.951) revealing no relationship between this polymorphism and psoriasis (Table 3). We did not identify between‐study heterogeneity (Table 3).
3.4. Sensitivity analysis and publication bias
We conducted sensitivity analyses to evaluate the stability of the overall effect by removing one study at a time and estimating the summary ORs for the remaining studies. The results remained essentially unchanged for the IL‐10 polymorphism rs1800896 and the IL‐1RN VNTR polymorphism (data not shown). Given that the meta‐analyses for the IL‐10 SNPs rs3021097 and rs1800872 were performed on a small number of small studies, sensitivity analysis was not carried out for these two SNPs. Begg's funnel plots (Figures 5 and 6) and Egger's test revealed no publication bias (p > 0.05 for each polymorphism).
Figure 5.
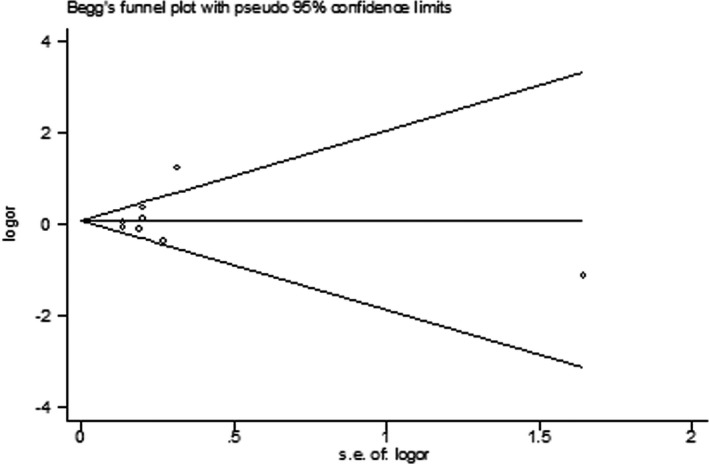
Begg's funnel plot for meta‐analysis of the interleukin‐1 receptor antagonist variable number tandem repeat polymorphism
Figure 6.
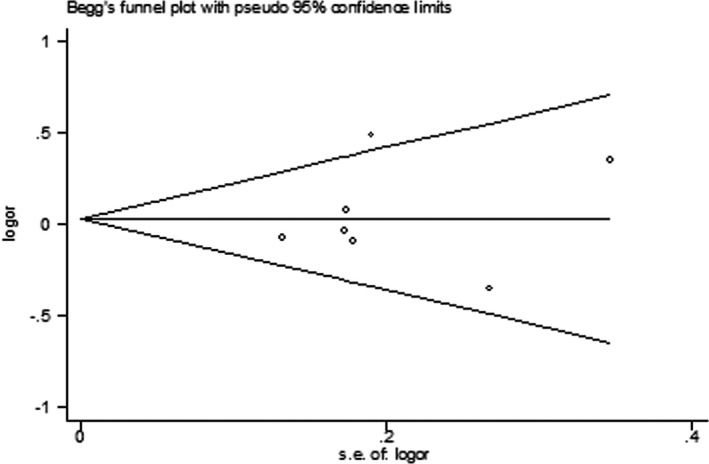
Begg's funnel plot for meta‐analysis of the interleukin‐10 polymorphism rs1800896
4. DISCUSSION
Interleukin‐1Ra and IL‐10 are critical negative regulators of the inflammatory response. Numerous studies have shown the effects of IL‐1Ra on blocking the activity of IL‐1α and IL‐1β in various in vitro and in vivo systems (Paolo & Shayakhmetov, 2016). IL‐1Ra knockout mice develop skin inflammation with histopathological features resembling human psoriasis (Nakajima et al., 2010; Shepherd et al., 2004), while administration of recombinant IL‐1Ra shows beneficial effects in patients with psoriasis (Viguier, Guigue, Pages, Smahi, & Bachelez, 2010). IL‐10 has a broader spectrum of anti‐inflammatory activities because it inhibits the production of a number of pro‐inflammatory cytokines, including interferon‐γ, IL‐8, IL‐6, IL‐1α, and IL‐1β. IL‐10 also up‐regulates the formation and release of anti‐inflammatory molecules. Treatment with recombinant IL‐10 has been shown to be effective in reducing the inflammatory reactions and disease severity in psoriasis patients (Asadullah et al., 1999; Reich et al., 2001). Because IL‐1ra and IL‐10 play an important role in controlling inflammation, it is possible that genetic variants in the IL‐1RN and IL‐10 genes may contribute to psoriasis susceptibility.
In this meta‐analysis, we systematically reviewed case–control studies on the association between psoriasis risk and genetic variants in the IL1‐RN and IL‐10 genes. The results of our meta‐analyses found no significant effect of the four individual polymorphisms (IL‐1RN VNTR polymorphism and IL‐10 SNPs rs1800896, rs3021097 and rs1800872) on psoriasis risk.
There is no previous meta‐analysis assessing the relation of the IL‐1RN VNTR polymorphism with psoriasis risk. Among the eight case–control studies on this polymorphism, conflicting results were reported. Three studies reported an association between this polymorphism and psoriasis (Liu et al., 1998; Peng & Wang, 1999; Tarlow et al., 1997), but the others did not find any associations (Chang et al., 2007; Li et al., 1999; Moorchung et al., 2015; Peddle et al., 2005; Reich et al., 2002). It is unclear what factors contribute to the conflicting results reported in these studies. Differences in genetic background, percentage of men, origin of controls, sample size, and environmental factors may be responsible for the inconsistent results. Meta‐analysis is a quantitative statistical analysis for synthesizing research results across different studies into an overall summary. Our study would benefit clinician by providing a summary of medical literature on the relationship of the IL‐1RN VNTR polymorphism with psoriasis. Based on a combined estimate of data from eight genetic studies involving 1,469 psoriasis patients and 1,262 control subjects, we found no association of the IL‐1RN VNTR polymorphism with psoriasis risk.
A previous meta‐analysis by Lee, Choi, Ji, and Song, (2012 assessed the IL‐10 SNPs and psoriasis risk in 2012. In their paper, Lee et al. did not reveal significant associations of rs1800896 and rs3021097 with psoriasis risk in the overall populations, but they identify a significant association between rs1800896 and psoriasis risk in Asian subjects. The SNP rs1800872 was not evaluated in their meta‐analysis. Lee et al. included two case–control studies for evaluating the rs1800896 polymorphism in Asians. In addition to the two studies Lee et al. used, we added a recently published Asian study with a large sample size (720 participants) into the pooled analysis, finding no significant association between rs1800896 and psoriasis in Asians. Given that the meta‐analysis by Lee et al. only included a small number of studies with small sample sizes for evaluating the relation of rs1800896 and psoriasis risk in Asians, the positive association they reported may be unreliable and needs to be confirmed by studies using larger sample size.
Finally, the limitations of our study must be discussed. First, haplotype analysis for the IL‐10 polymorphisms was not performed because of limited number of studies (n = 2) and discrepancy in study methods (Al‐Heresh et al., 2002; Baran, Szepietowski, Mazur, & Baran, 2008). It was noted that the two studies evaluating IL‐10 haplotype and psoriasis risk did not reported a significant association. Second, owing to low sample size of studies, the association of the IL‐1RN VNTR polymorphism and IL‐10 SNPs with psoriasis subtypes was not taken into account in our meta‐analysis. Third, there were discrepancies in the presentation of published data for the IL‐1RN VNTR polymorphism. It is recommended that further studies should provide data for both genotype and allelotype frequencies of the polymorphism.
In conclusion, current published studies fail to support the hypothesis that the IL‐1RN VNTR polymorphism and three common IL‐10 SNPs rs1800896, rs3021097, and rs1800872 are associated with psoriasis risk.
CONFLICTS OF INTEREST
The authors declare no conflict of interest.
Qiao J, Jia Q‐N, Jin H‐Z. Lack of association of the IL‐1RN and IL‐10 polymorphisms with risk of psoriasis: A meta‐analysis. Mol Genet Genomic Med. 2019;7:e512 10.1002/mgg3.512
REFERENCES
- Al‐Heresh, A. M. , Proctor, J. , Jones, S. M. , Dixey, J. , Cox, B. , Welsh, K. , & McHugh, N. (2002). Tumour necrosis factor‐alpha polymorphism and the HLA‐Cw*0602 allele in psoriatic arthritis. Rheumatology, 41(5), 525–530. 10.1093/rheumatology/41.5.525 [DOI] [PubMed] [Google Scholar]
- Asadullah, K. , Docke, W. D. , Ebeling, M. , Friedrich, M. , Belbe, G. , Audring, H. , … Sterry, W. (1999). Interleukin 10 treatment of psoriasis: Clinical results of a phase 2 trial. Archives of Dermatology, 135(2), 187–192. 10.1001/archderm.135.2.187 [DOI] [PubMed] [Google Scholar]
- Baran, W. , Szepietowski, J. C. , Mazur, G. , & Baran, E. (2008). IL‐6 and IL‐10 promoter gene polymorphisms in psoriasis vulgaris. Acta Dermato‐Venereologica, 88(2), 113–116. 10.2340/00015555-0427 [DOI] [PubMed] [Google Scholar]
- Bid, H. K. , Manchanda, P. K. , & Mittal, R. D. (2006). Association of interleukin‐1Ra gene polymorphism in patients with bladder cancer: Case control study from North India. Urology, 67(5), 1099–1104. 10.1016/j.urology.2005.11.032 [DOI] [PubMed] [Google Scholar]
- Boehncke, W. H. , & Schon, M. P. (2015). Psoriasis. Lancet, 386(9997), 983–994. 10.1016/S0140-6736(14)61909-7 [DOI] [PubMed] [Google Scholar]
- Chang, Y. T. , Chou, C. T. , Yu, C. W. , Lin, M. W. , Shiao, Y. M. , Chen, C. C. , … Tsai, S. F. (2007). Cytokine gene polymorphisms in Chinese patients with psoriasis. The British Journal of Dermatology, 156(5), 899–905. 10.1111/j.1365-2133.2007.07820.x [DOI] [PubMed] [Google Scholar]
- Cheng, J. , Tu, Y. , Li, J. , Huang, C. , Liu, Z. , & Liu, D. A study on the expression of interleukin (IL)‐10 and IL‐12 P35, P40 mRNA in the psoriatic lesions. Journal of Tongji Medical University, 2001; 21(1), 86–88. [DOI] [PubMed] [Google Scholar]
- Craven, N. M. , Jackson, C. W. , Kirby, B. , Perrey, C. , Pravica, V. , Hutchinson, I. V. , & Griffiths, C. E. (2001). Cytokine gene polymorphisms in psoriasis. The British Journal of Dermatology, 144(4), 849–853. 10.1046/j.1365-2133.2001.04143.x [DOI] [PubMed] [Google Scholar]
- Debets, R. , Hegmans, J. P. , Croughs, P. , Troost, R. J. , Prins, J. B. , Benner, R. , & Prens, E. P. (1997). The IL‐1 system in psoriatic skin: IL‐1 antagonist sphere of influence in lesional psoriatic epidermis. Journal of Immunology, 158(6), 2955–2963. [PubMed] [Google Scholar]
- Di Paolo, N. C. , & Shayakhmetov, D. M. (2016). Interleukin 1alpha and the inflammatory process. NatureImmunology, 17(8), 906–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontoura, I. C. , Trombone, A. P. , Almeida, L. P. , Lorenzi, J. C. , Rossetti, R. A. , Malardo, T. , … Coelho‐Castelo, A. A. M. (2015). B cells expressing IL‐10 mRNA modulate memory T cells after DNA‐Hsp65 immunization. Brazilian Journal of Medical and Biological Research, 48(12), 1095–1100. 10.1590/1414-431X20154409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi, M. , Yanaba, K. , Umezawa, Y. , Yoshihara, Y. , Kikuchi, S. , Ishiuji, Y. , … Nakagawa, H. (2017). Corrigendum to “IL‐10‐producing regulatory B cells are decreased in patients with psoriasis” [J. Dermatol. Sci. 81 (2016) 93–100]. Journal of Dermatological Science, 86(1), 93–100. 10.1016/j.jdermsci.2017.01.010 [DOI] [PubMed] [Google Scholar]
- Indhumathi, S. , Rajappa, M. , Chandrashekar, L. , Ananthanarayanan, P. H. , Thappa, D. M. , & Negi, V. S. (2017). T helper‐2 cytokine/regulatory T‐cell gene polymorphisms and their relation with risk of psoriasis in a South Indian Tamil cohort. Human Immunology, 78(2), 209–215. 10.1016/j.humimm.2016.12.006 [DOI] [PubMed] [Google Scholar]
- Karam, R. A. , Zidan, H. E. , & Khater, M. H. (2014). Polymorphisms in the TNF‐alpha and IL‐10 gene promoters and risk of psoriasis and correlation with disease severity. Cytokine, 66(2), 101–105. [DOI] [PubMed] [Google Scholar]
- Kim, W. B. , Jerome, D. , & Yeung, J. (2017). Diagnosis and management of psoriasis. Canadian Family Physician Medecin De Famille Canadien, 63(4), 278–285. [PMC free article] [PubMed] [Google Scholar]
- Kristensen, M. , Deleuran, B. , Eedy, D. J. , Feldmann, M. , Breathnach, S. M. , & Brennan, F. M. (1992). Distribution of interleukin 1 receptor antagonist protein (IRAP), interleukin 1 receptor, and interleukin 1 alpha in normal and psoriatic skin. Decreased expression of IRAP in psoriatic lesional epidermis. The British Journal of Dermatology, 127(4), 305–311. [DOI] [PubMed] [Google Scholar]
- Lee, Y. H. , Choi, S. J. , Ji, J. D. , & Song, G. G. (2012). Associations between interleukin‐10 polymorphisms and susceptibility to psoriasis: A meta‐analysis. Inflammation, 61(7), 657–663. 10.1007/s00011-012-0458-2 [DOI] [PubMed] [Google Scholar]
- Li, G. , Dong, H. , Zhu, X. , Yu, S. , & Wang, S. (1999). Preliminary exploration of the relationship between IL 1ra intron 2 polymorphism and psoriasis. Journal of Clinical Dermatology. [Google Scholar]
- Liu, Z. F. , Zhang, J. M. , Yang, Q. , & Xu, T. H. (1998). Association between interleukin‐1 receptor antagonist gene polymorphism and psoriasis. National Medical Journal of China, 78, 525–526. [Google Scholar]
- Mahil, S. K. , Capon, F. , & Barker, J. N. (2016). Update on psoriasis immunopathogenesis and targeted immunotherapy. Seminars in Immunopathology, 38(1), 11–27. 10.1007/s00281-015-0539-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantel, N. , & Haenszel, W. (1959). Statistical aspects of the analysis of data from retrospective studies of disease. Journal of the National Cancer Institute, 22(4), 719–748. 10.1093/jnci/22.4.719 [DOI] [PubMed] [Google Scholar]
- Mistry, A. , Savic, S. , & van der Hilst, J. C. H. (2017). Interleukin‐1 Blockade: An Update on Emerging Indications. BioDrugs: Clinical Immunotherapeutics, Biopharmaceuticals and Gene Therapy, 31(3), 207–221. 10.1007/s40259-017-0224-7 [DOI] [PubMed] [Google Scholar]
- Moorchung, N. , Vasudevan, B. , Chatterjee, M. , Mani, N. S. , & Grewal, R. S. (2015). Interleukin‐1 gene polymorphisms and their relation with NFkappaB expression and histopathological features in psoriasis. Indian Journal of Dermatology, 60(5), 432–438. 10.4103/0019-5154.159630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima, A. , Matsuki, T. , Komine, M. , Asahina, A. , Horai, R. , Nakae, S. , … Iwakura, Y. (2010). TNF, but not IL‐6 and IL‐17, is crucial for the development of T cell‐independent psoriasis‐like dermatitis in Il1rn‐/‐ mice. Journal of Immunology, 185(3), 1887–1893. 10.4049/jimmunol.1001227 [DOI] [PubMed] [Google Scholar]
- Peddle, L. , Butt, C. , Snelgrove, T. , & Rahman, P. (2005). Interleukin (IL) 1alpha, IL1beta, IL receptor antagonist, and IL10 polymorphisms in psoriatic arthritis. Annals of the Rheumatic Diseases, 64(7), 1093–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, X. B. , & Wang, Z. F. (1999). Study of the association between psoriasis and interleukin‐1 receptor antagonist gene polymorphism. Journal of Immunology, 15, 188–189. [Google Scholar]
- Reich, K. , Garbe, C. , Blaschke, V. , Maurer, C. , Middel, P. , Westphal, G. , … Neumann, C. (2001). Response of psoriasis to interleukin‐10 is associated with suppression of cutaneous type 1 inflammation, downregulation of the epidermal interleukin‐8/CXCR34 pathway and normalization of keratinocyte maturation. The Journal of Investigative Dermatology, 116(2), 319–329. 10.1046/j.1523-1747.2001.01248.x [DOI] [PubMed] [Google Scholar]
- Reich, K. , Mossner, R. , Konig, I. R. , Westphal, G. , Ziegler, A. , & Neumann, C. (2002). Promoter polymorphisms of the genes encoding tumor necrosis factor‐alpha and interleukin‐1beta are associated with different subtypes of psoriasis characterized by early and late disease onset. The Journal of Investigative Dermatology, 118(1), 155–163. [DOI] [PubMed] [Google Scholar]
- Reich, K. , Westphal, G. , Schulz, T. , Muller, M. , Zipprich, S. , Fuchs, T. , … Neumann, C. (1999). Combined analysis of polymorphisms of the tumor necrosis factor‐alpha and interleukin‐10 promoter regions and polymorphic xenobiotic metabolizing enzymes in psoriasis. The Journal of Investigative Dermatology, 113(2), 214–220. [DOI] [PubMed] [Google Scholar]
- Renne, J. , Schafer, V. , Werfel, T. , & Wittmann, M. (2010). Interleukin‐1 from epithelial cells fosters T cell‐dependent skin inflammation. The British Journal of Dermatology, 162(6), 1198–1205. 10.1111/j.1365-2133.2010.09662.x [DOI] [PubMed] [Google Scholar]
- Rutz, S. , & Ouyang, W. (2016). Regulation of interleukin‐10 expression. Advances in Experimental Medicine and Biology, 941, 89–116. [DOI] [PubMed] [Google Scholar]
- SanGiovanni, J. P. , Berkey, C. S. , Dwyer, J. T. , & Colditz, G. A. (2000). Dietary essential fatty acids, long‐chain polyunsaturated fatty acids, and visual resolution acuity in healthy fullterm infants: A systematic review. Early Human Development, 57(3), 165–188. 10.1016/S0378-3782(00)00050-5 [DOI] [PubMed] [Google Scholar]
- Shepherd, J. , Little, M. C. , & Nicklin, M. J. (2004). Psoriasis‐like cutaneous inflammation in mice lacking interleukin‐1 receptor antagonist. The Journal of Investigative Dermatology, 122(3), 665–669. 10.1111/j.0022-202X.2004.22305.x [DOI] [PubMed] [Google Scholar]
- Tarlow, J. K. , Cork, M. J. , Clay, F. E. , Schmitt‐Egenolf, M. , Crane, A. M. , Stierle, C. , … Duff, G. V. (1997). Association between interleukin‐1 receptor antagonist (IL‐1ra) gene polymorphism and early and late‐onset psoriasis. The British Journal of Dermatology, 136(1), 147–148. 10.1111/j.1365-2133.1997.tb08779.x [DOI] [PubMed] [Google Scholar]
- Viguier, M. , Guigue, P. , Pages, C. , Smahi, A. , & Bachelez, H. (2010). Successful treatment of generalized pustular psoriasis with the interleukin‐1‐receptor antagonist Anakinra: Lack of correlation with IL1RN mutations. Annals of Internal Medicine, 153(1), 66–67. 10.7326/0003-4819-153-1-201007060-00030 [DOI] [PubMed] [Google Scholar]
- Wongpiyabovorn, J. , Hirankarn, N. , Ruchusatsawat, K. , Yooyongsatit, S. , Asawanonda, P. , & Poovorawan, Y. (2008). Association of the interleukin‐10 distal promoter (‐2763A/C) polymorphism with late‐onset psoriasis. Clinical and Experimental Dermatology, 33(2), 186–189. 10.1111/j.1365-2230.2007.02628.x [DOI] [PubMed] [Google Scholar]
- Yanaba, K. , Kamata, M. , Ishiura, N. , Shibata, S. , Asano, Y. , Tada, Y. , … Sato, S. (2013). Regulatory B cells suppress imiquimod‐induced, psoriasis‐like skin inflammation. Journal of Leukocyte Biology, 94(4), 563–573. 10.1189/jlb.1112562 [DOI] [PMC free article] [PubMed] [Google Scholar]


